QTL detection for milk production traits in goats using a longitudinal model
Summary
Eight paternal half-sib families were used to identify chromosomal regions associated with variation in the lactation curves of dairy goats. DNA samples from 162 animals were amplified by PCR for 37 microsatellite markers, from Capra hircus autosomes CHI3, CHI6, CHI14 and CHI20. Milk samples were collected during 6 years, and there were 897 records for milk yield (MY) and 814 for fat (FP) and protein percentage (PP). The analysis was conducted in two stages. First, a random regression model with several fixed effects was fitted to describe the lactation function, using a scale (α) plus four shape parameters: β and γ, both associated with a decrease in the slope of the curve, and δ and φ that are related to the increase in slope. Predictions of α, β, γ, δ and φ were regressed using an interval mapping model, and F-tests were used to test for quantitative trait loci (QTL) effects. Significant (p < 0.05) QTLs were found for: (i) MY: CHI6 at 70–80 cM for all parameters; CHI14 at 14 cM for δ and φ; (ii) FP: CHI14, at 63 cM was associated with β; CHI20, at 72 cM, showed association with α; (iii) PP: chromosomal regions associated with β were found at 59 cM in CHI3 and at 55 cM in CHI20 with α and γ. Analyses using more families and more animals will be useful to confirm or to reject these findings.
Introduction
Most studies of quantitative trait loci (QTL) detection for milk production traits have been carried out in cattle, using mostly aggregated data (Zhang et al. 1998; Heyen et al. 1999; Plante et al. 2001; Viitala et al. 2003; Ashwell et al. 2004). Genetic associations between molecular markers and economically important traits in goats have been reported by Cano et al. (2007) and Marrube et al. (2007). However, the only well-documented genetic association with dairy traits in goats is the one related with the highly variable alpha S1-casein polymorphisms (Grosclaude et al. 1994; Adnoy et al. 2003; Manfredi 2003; Suárez 2004; Sacchi et al. 2005). Analyses on QTLs affecting milk traits of dairy goats are lacking.
In searching for QTLs in goats, one may look at those significant associations found between milk production traits and genetic markers in cattle (reviewed by Khatkar et al. 2004), and take advantage of the homology between the genetic maps of the cow and the goat. Moreover, Rodriguez-Zas et al. (2002) measured the association between chromosomal regions and the scale and shape that describe the lactation curve in dairy cattle using a two-stage procedure: a random regression model to predict the elements of the lactation function on an animal basis, followed by a regression interval mapping using the predictions obtained in the first stage of the analysis. The goal of this research was to identify chromosomal regions associated with variation in the lactation function of goats using the two-stage procedure employed by Rodriguez-Zas et al. (2002).
Material and methods
Description of phenotypic data
The population used in the study was established in 1998 at the Regional Experimental Center of Leales – INTA, in the province of Tucumán, Argentina. Phenotypes recorded were milk yield (MY), fat percentage (FP) and protein percentage (PP) from 212 female goats. Milk samples were collected at the morning milking, at each of the two kidding seasons (fall-winter and spring-summer) during 6 years (1999–2004). Each goat was sampled for FP and PP once a month, either five or six times per lactation. Records of daily MY for a given test day were the averages of an entire week, with sampling taking place every month but on an irregular basis (up to 3 weeks a month). Lactations with three or fewer records were deleted, and the lactation stage ranged from 3 to 321 days. The total number of observations were 897 for MY, and 814 for FP and PP. Averages (standard deviations) across all families were 0.879 kg (SD 0.563) for MY, 4.382% (SD 1.864) for FP and 4.065% (SD 0.757) for PP.
Genotypic data
Eight paternal half-sib families composed of 87 young females and 75 older goats were used for QTL detection. Whereas six families were purebred Criollo goats, the two other families consisted of Saanen by Criollo crosses. The Criollo goat is a local dairy-like breed living in harsh environments. The breed has remained almost unselected since its introduction from Spain nearly 500 years ago (Rodero et al. 1992). The number of females recorded for MY and FP or PP, and the number of genotyped goats per family are displayed in Table 1.
| Families | Breed composition | Number of recorded goats | Total number of records | Number of genotyped goats | |
|---|---|---|---|---|---|
| MY | Contents | ||||
| 1 | Criollo | 30 | 114 | 112 | 25 |
| 2 | Criollo | 22 | 92 | 83 | 16 |
| 3 | Criollo | 26 | 107 | 91 | 25 |
| 4 | Criollo | 23 | 99 | 93 | 18 |
| 5 | Criollo | 19 | 78 | 67 | 15 |
| 6 | Criollo | 16 | 46 | 34 | 7 |
| 7 | Saanen cross | 20 | 85 | 74 | 14 |
| 8 | Saanen cross | 56 | 276 | 260 | 42 |
| Total | – | 212 | 897 | 814 | 162 |
- MY, milk yield.
DNA was extracted from whole blood using the technique by Madisen et al. (1987). PCR reactions were carried out using fluorescence or γ32P ATP-primers labels as previously described by Cano et al. (2007), and the PCR products were separated by electrophoresis in denaturing polyacrylamide gels, and visualized in an automatic sequencer or in autoradiography. We selected four Capra hircus autosomes (CHI), based on the homologies between the genetic maps of cattle and goats and the significant associations summarized by Khatkar et al. (2004) in Bos taurus autosome (BTA) 3, 6, 14 and 20.
A total of 37 microsatellite markers (MS) were selected from the web genetic map resources (http://locus.jouy.inra.fr/cgi-bin/lgbc/mapping/common/intro2.pl?BASE=goat, http://locus.jouy.inra.fr/cgi-bin/lgbc/mapping/common/intro2.pl?BASE=cattle, http://www.animalgenome.org/cattle/maps/db.html and http://bioinformatics.roslin.ac.uk). The linkage maps were used as a guide for intermarker distance, and the microsatellites were positioned and ordered on the chromosomes with the cri-map 2.4 program (Green et al. 1990). A summary of the informative markers used in the study across the four chromosomes are shown in Table 2. The average number of informative markers by autosome was 6 (ranging from 4 to 8), with an average spacing of 19.22 cM. The highest proportion of heterozygous sires averaged over all markers by chromosome was in CHI6. The average heterozygosity was 60.72%, and the average number of alleles by marker across families was 6.7. The markers showing the highest polymorphic information content (PIC) were microsatellites BM4621, BM143 and CSN, with PIC values of 0.798, 0.789 and 0.763, respectively, and located in CHI6.
| CHI | Number of markers | PIC | Proportion of heterozygous siresa | Marker distances (cM) |
|---|---|---|---|---|
| 3 | 4 | 0.604 | 0.50 | INRA006 (0.0), McM58 (23.4), INRA023 (29.7), HUJ177 (17.5) |
| 6 | 5 | 0.665 | 0.62 | BM1329 (0.0), BM143 (31.7), BM4621 (34.4), BM415 (23.3), CSN (42.3) |
| 14 | 8 | 0.511 | 0.58 | ETH225 (0.0), ILSTS011 (9.4), RM011 (26.8), CSSM66 (17.8), CSSM36 (9.1), BMC1207 (20.4), BM302 (36.5), BM4513 (38.8) |
| 20 | 7 | 0.516 | 0.43 | TGLA304 (0.0), TGLA443 (6.4), BM4107 (17.2), INRA036 (17.6), OarHH62 (13.2), ILSTS072 (10.9), BMS1719 (7.1) |
- CHI, Capra hircus autosome; PIC, polymorphic information content.
- aProportion of heterozygous sires averaged over all markers by chromosome.
Statistical analysis
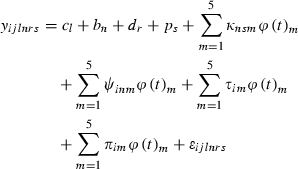 (1)
(1)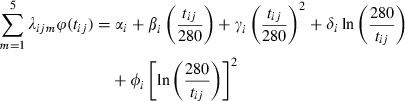
In the second stage, we performed a QTL analysis using the half-sib regression interval mapping method of Knott et al. (1996), with the software QTL Express (Seaton et al. 2002). The test statistics were computed every centiMorgan over the mapped chromosome. F-statistic thresholds for chromosome-wise level were calculated from 10 000 permutations (Churchill & Doerge 1994). Families that displayed the highest evidence for a QTL at the location in the across-family analysis were taken from the QTL-express output (Knott et al. 1996).
Results and discussion
The estimated heritability for MY ranged from 0.142 to 0.593, and the average estimated over the whole lactation period was 0.343. These values were in agreement to those reported in the literature for different goat breeds, and with estimates obtained from either single or multiple trait models. Our results seem to be slightly lower than those of Weppert & Hayes (2004) for Nubian, Alpine, Saanen and Toggenburg goats, when maternal effects were included in the analysis. The estimated heritability of MY from a model without maternal effects was equal to 0.19 (Weppert & Hayes 2004). Similar estimates of heritability to the values found in the current study were reported for South African Saanen goats (0.30, Muller et al. 2002), and for Alpine and Saanen females (0.23, Clément et al. 2002).
The average heritability estimate of FP was equal to 0.092 (ranging from 0.093 to 0.141), and the average heritability estimate of PP was 0.160 (ranging from 0.007 to 0.515). These values were lower than those obtained and reviewed by Muller et al. (2002) using data from several goat breeds: the range of FP was 0.160–0.540 (Spain, Alpine, Saanen and Toggenburg breeds) and the range of PP was 0.250–0.620 (Spain, Greece and Saanen goats).
For the QTL analysis, all significant F-statistics and their p-values for the three traits obtained from the regression interval mapping method of Knott et al. (1996) are displayed in Table 3.
| Autosome | Traits | λa | Map position (cM) | F-statistics (p-values) | Family |
|---|---|---|---|---|---|
| 3 | PP | β | 59 | 2.99 (0.01205) | 5/7 |
| 6 | MY | α | 75 | 4.35 (0.00016) | 7 |
| 6 | MY | β | 78 | 4.89 (0.00004) | 7 |
| 6 | MY | γ | 75 | 4.38 (0.00016) | 7 |
| 6 | MY | δ | 71 | 3.48 (0.00146) | 7 |
| 6 | MY | φ | 70 | 3.20 (0.00293) | 7 |
| 14 | MY | δ | 14 | 2.35 (0.04062) | 7 |
| 14 | MY | φ | 14 | 2.42 (0.03559) | 7 |
| 14 | FP | β | 63 | 2.63 (0.01300) | 6 |
| 20 | PP | α | 55 | 4.49 (0.00056) | 5 |
| 20 | PP | γ | 55 | 4.56 (0.00049) | 5 |
| 20 | FP | α | 72 | 4.71 (0.00034) | 5 |
- MY, milk yield; FP, fat percentage; PP, protein percentage.
- aParameters of the lactation function. α, scale parameter; β and γ, decreasing slope; δ and φ, increasing slope.
Milk yield
Seven tests were significant for at least one parameter. In CHI6, we detected a significant effect for all parameters in the interval flanked for the MS BM4621 and BM415 (from 70 to 78 cM). One chromosomal region was associated with δ and φ on CHI14. Several studies based on cumulative single records in dairy cattle detected the presence of QTL at the genome-wise and suggestive thresholds on BTA3, in the interval from 16 to 32 cM (Heyen et al. 1999), at 39 cM (Plante et al. 2001), and at 40 cM (Vandervoort & Jansen 2002). On the other hand, neither Viitala et al. (2003) nor Ashwell et al. (2004), reported evidence of a QTL for MY on BTA3. Rodriguez-Zas et al. (2002) using test-day milk records reported a significant association between marker MS BL41 (32 cM) and the parameter that describes the shape of the function at the beginning of the lactation. Additionally, these workers reported significant associations between the scale parameter and two chromosomal regions of BTA3 that were located in the centromere from 0 to 36 cM and in the telomere from 91 to 113 cM (close to MS HUJI177 at 100 cM and MS BR4502 at 113 cM).
An association with increasing slope parameters δ and φ was found at 70 and 71 cM, respectively, on CHI6. Also at 75 cM, the scale parameter (α) and the descriptor of the shape at the end of the lactation (γ) were found to be significant. Another association with the remained descriptor of the decreasing slope (β) was detected at 78 cM. The correlation between the estimates of α, γ, δ and φ was high (0.922–0.998). Similarly, the correlation between β and the other parameters ranged from 0.652 to 0.758. Such correlations may suggest a QTL with pleiotropic effects on these parameters. Rodriguez-Zas et al. (2002) detected a putative QTL between 0 and 21 cM in dairy cattle that affected the scale parameter for MY. They also reported the finding of another QTL affecting the shape parameters for MY, which is located in the region from 108 to 129 cM. In dairy cattle, Zhang et al. (1998) reported a putative QTL in the interval between 30 to 50 cM on BTA6 (between MS BM1329 and BM143).
In the goat, the casein gene cluster has been mapped to the distal region of CHI6 (Grosclaude et al. 1994), and is composed of four genes (αS1-casein, αS2-casein, β-casein, and κ-casein). Moioli et al. (2007) reviewed the several associations between casein genes, and dairy traits of goats have been reported in the literature. This is especially so for the αS1 gene, which displays a higher level of polymorphism than the one observed in the bovine, and has been related to fat and protein contents (Grosclaude et al. 1994; Adnoy et al. 2003; Manfredi 2003; Sacchi et al. 2005). However, the effect of αS1 casein alleles on MY does not seem to be important (Moioli et al. 2007). In the current research, we used αS1 casein gene as a marker gene (MS CSN) and did not find association to MY. A QTL on chromosome 6 of cattle located closer to the casein gene cluster was reported by Khatkar et al. (2004).
Moving to CHI14, there was evidence for a QTL at 14 cM (the interval flanked by ILSTS011 and RM011) related to δ and φ. This result suggests that there may be a QTL with pleiotropic effects on both parameters. It may also be the case that, due to the high correlation (0.996) between the estimates of those parameters, one of the associations observed may be a false positive. Similarly, Rodriguez-Zas et al. (2002) reported an association between the chromosomal region at about 13 cM on BTA14 (marker CSSM66), and the shape parameters that describe the changes in milk yield during mid and late lactation. However, in dairy cattle a QTL affecting MY was seemingly associated with BTA14 by Khatkar et al. (2004). Several studies in cattle reported QTLs for MY on BTA20. The chromosomal regions in the bovine were at 21 cM (Plante et al. 2001), 82 cM (Viitala et al. 2003) and 68 cM (Ashwell et al. 2004). In the current study, no microsatellite marker on CHI20 was associated with some lactation descriptors.
Fat percentage
Although several studies in dairy cattle reported a putative QTL on BTA3 and BTA6 affecting FP (Khatkar et al. 2004), we did not find evidence for the effects of a chromosomal region in either CHI3 or CHI6, at the chromosomal-wise threshold. However, for FP we detected significant effects from CHI14 and CHI20.
When analyzing CHI14, one chromosomal region at 63 cM (between CSSM66 and CSSM36) associated with variation of the parameter β was found significant. Conversely, analyses with dairy cattle found strong evidence for a putative QTL affecting FP near the centromere of BTA14, being the MS CSSM66 the nearest marker (Coppieters et al. 1998; Zhang et al. 1998; Heyen et al. 1999; Ashwell et al. 2004). A QTL proximal to the centromere on BTA14 with an effect on FP has consistently been reported (Grisart et al. 2002 and Winter et al. 2002), and the mutation underlying this QTL has been identified (Winter et al. 2002) as the K232A substitution in exon VIII of acylCoA/diacylglycerol acyltransferase 1 enzyme (DGAT1). This enzyme is considered to be of importance in controlling the synthesis rate of triglycerides in adipocytes. Nevertheless, no associations with several microsatellite markers on BTA14 were found by Rodriguez-Zas et al. (2002) using a longitudinal mapping model. Comparing the goat and bovine linkage maps, CHI14 shows a partial homology with BTA14 from MS CSSM66 to the telomere. The centromeric region of CHI14 flanked between MS ETH225 and MS BM757, corresponds to the same region of chromosome 9 in cattle. Therefore, it is likely that an association with FP could be found near the centromeric end of CHI9.
At 72 cM on CHI20, where the nearest marker is BMS1719, we detected an association with α (p < 0.00002). Many additional QTL with significant effects on FP and FY has been reported for chromosome 20 in dairy cattle (Khatkar et al. 2004).
Protein percentage
Chromosomal regions associated with β for PP were found in CHI3. For this chromosome and close to MS INRA023 (at 59 cM), we detected a chromosomal region affecting β. A QTL for PP located in an area of about 40 cM in BTA3 was reported in several studies (Khatkar et al. 2004).
We did not find significant associations among chromosomal regions of CHI6 and any parameter when looking at PP. Nevertheless, many studies in dairy cattle have detected the presence of a QTL close to MS BM143 in BTA6 that is related to PP, and the marker position agrees in all these studies (Spelman et al. 1996; Zhang et al. 1998; Ron et al. 2001; Viitala et al. 2003). Using longitudinal phenotypic data, Rodriguez-Zas et al. (2002) found significant association between MS BM143 and the scale and the shape parameters at middle and late lactation for PP in dairy cows. Recently, Schnabel et al. (2005) performed a fine-mapping study of BTA6 of dairy cattle and identified the osteopontin (OPN) gene as an ideal functional candidate gene for a QTL very close to BM143. The OPN is a secreted glycoprotein and its expression in the murine mammary gland depends on the stage of postnatal development, which in turn suggests a role for OPN in mammary involution.
Lactation patterns
We use the expression longitudinal mapping model to refer to those statistical models for mapping QTLs of a longitudinal (or functional value) trait. The ability of the model used in the current study to detect associations between markers and lactation stages may contribute to explain the different lactation patterns among individuals.
Although breeders do not usually breed for lactation shape, some traits such as persistence that are described by lactation curve parameters are economically relevant. We have looked at MY persistency. However, we did not find any significant association with any of the markers studied here.
General
Although the function of Ali & Schaeffer (1987) may suffer from high correlations among the five predicted parameters, Macciota et al. (2005) observed that certain combinations in the signs of the parameters were more prone to a high correlation than others. For the correlation patterns observed among the five predicted parameters ranged from 0.658 (correlation between β and φ) to 0.998 (correlation between α and γ). The function of Ali & Schaeffer (1987) is flexible as it allows accounting for differences in the rate of change in milk along the entire lactation, thus displaying a large number of different curves as described by Macciota et al. (2005).
A limitation of this study is the less number of animals involved in the analysis, which was coupled with a limited size of the half-sib families involved. Clearly, these results have low statistical power. Even if the results are not conclusive, they provide, nevertheless, a general idea of potential QTL locations in Criollo and Creole cross goats. Analyses using more families and more animals will be useful to confirm or to reject our findings.
To our knowledge, this is the first report in goats searching for chromosomal regions associated with variation in the lactation curve. Nine map positions were identified as affecting any parameter of the lactation function of dairy goats: one in CHI3, four in CHI6, two in CHI14 and two in CHI20. Some of these results were consistent with QTLs found in dairy cattle while using either aggregate lactation records, or longitudinal-linkage analysis.
Acknowledgements
The authors thank SeCyT-ANPCyT, Argentina, through the National Project BID1728 PICTO12968, Universidad de Buenos Aires, through grants UBACyT G018-Program 2004–2007 and CONICET PIP 5338 2006–2007; La Serenísima S.A., Argentina, for their cooperation in supplying compositional milk data. We would also like to thank two anonymous reviewers for their constructive comments and suggestions that greatly enhanced the presentation.




