Direct genotyping of the poplar leaf rust fungus, Melampsora medusae f. sp. deltoidae, using codominant PCR-SSCP markers
Génotypage direct de la rouille des feuilles du peuplier, Melampsora medusae f. sp. deltoidae, par l'utilisation de marqueurs codominants PCR-SSCP
Direktes Genotyping des Pappelblattrostes Melampsora medusae f.sp. deltoidae mit codominanten PCR-SSCP-Markern
Summary
enTwo anonymous DNA markers that are revealed by single-strand conformational polymorphism (SSCP) analysis were developed for detection of polymorphisms in Melampsora medusae f. sp. deltoidae (Mmd). Mono-uredinial isolates of Mmd were first obtained, DNA was extracted from urediniospores and random amplified polymorphic DNA (RAPD) products of eight mono-uredinial isolates were separated on a SSCP gel to identify differences among them. Bands representing putative polymorphic loci among the eight isolates tested were excised from the SSCP gel and re-amplified by polymerase chain reaction (PCR), and then cloned and sequenced. A primer pair was designed to amplify a DNA fragment of a size suitable for SSCP analysis (<600 bp) for two out of three DNA fragments sequenced. Each set of primers amplified a PCR product for all eight isolates that were initially used to generate them and the resulting PCR products were analysed by SSCP. Polymorphisms among isolates were identified for both putative loci. The two primer pairs amplified a PCR product of the expected size on an additional 32 mono-uredinial isolates of Mmd tested. From the overall 40 mono-uredinial isolates tested, 5 and 11 alleles were detected, and 12 and 34 isolates showed to be heterozygous, as indicated by the presence of more than two bands on the SSCP gel, at loci A and B, respectively. The primer pairs were tested for specificity against 106 fungal isolates belonging to various taxa, including other rusts, and against DNA extracted from greenhouse-grown healthy poplar leaves. DNA amplification products of the expected size were obtained only when Mmd DNA was present. Optimization of PCR conditions with these two primer pairs allowed genotyping directly from single uredinia extracted from infected leaves, thus alleviating the need to culture the fungus to characterize individuals, hence making it possible to process large numbers of samples for population studies.
Résumé
frDeux marqueurs génétiques anonymes, révélés par analyse SSCP (Single-Strand Conformational Polymorphism) ont été développés afin de détecter des polymorphismes génétiques chez le Melampsora medusae f. sp. deltoidae (Mmd). Dans un premier temps, des isolats mono-urédiniaux ont été obtenus, puis l'ADN a été extrait à partir des urédiniospores, les produits d'amplification RAPD (Random Amplified Polymorphic DNA) ont été générés à partir de huit de ces isolats mono-urédiniaux et les résultats d'amplification ont par la suite été séparés sur gel SSCP afin d'identifier des polymorphismes entre les isolats. Les bandes sur gel SSCP représentant des loci polymorphiques putatifs entre les isolats ont été prélevées du gel, ré-amplifiées par la technique d'amplification PCR (Polymerase Chain Reaction), clonées, puis séquencées. Pour deux fragments d'ADN séquencés sur un total de trois, une paire d'amorces a été développée afin de permettre l'amplification d'un fragment de taille adéquate pour analyse SSCP (<600 pb). Chaque paire d'amorces a produit un signal d'amplification positif pour chacun des huit isolats à l'origine de ces nouvelles amorces; les produits PCR ont ensuite été analysés par la technique SSCP. Les deux loci putatifs ont révélé des polymorphismes génétiques entre les isolats. Les deux paires d'amorces ont produit un fragment d'amplification de la taille attendue pour chacun des 32 isolats mono-urédiniaux supplémentaires testés. Des 40 isolats testés, 5 et 11 allèles ont été détectés, alors que 12 et 34 isolats se sont révélés hétérozygotes (tel qu'indiqué par la présence de plus de deux bandes sur gel SSCP) pour les loci A et B, respectivement. La spécificité des deux paires d'amorces a été testée à partir de 106 isolats fongiques appartenant à différents groupes taxonomiques, incluant d'autres rouilles, de même qu’à partir de l'ADN extrait de feuilles de peupliers cultivés en serre. Un signal d'amplification positif n'a été obtenu qu'en présence d'ADN du Mmd. Les conditions d'amplification PCR ont été optimisées pour les deux paires d'amorces développées afin de permettre le génotypage directement à partir d'urédinies individuelles prélevées sur des feuilles de peuplier infectées. La possibilité de génotyper directement des urédinies individuelles permet d’éviter l'obligation de cultiver le champignon pour génotyper les individus, ce qui représente un avantage important des marqueurs génétiques développés ici, puisqu'il devient dès lors possible de traiter un grand nombre d’échantillons lors de la réalisation d’études de populations.
Zusammenfassung
deZum Nachweis von Polymorphismen bei Melampsora medusae f. sp. deltoidae wurden zwei anonyme DNA Marker aus einer SSCP-Analyse entwickelt. Zunächst wurden Isolate aus einzelnen Uredinien gewonnen, die DNA wurde aus den Uredosporen extrahiert und polymorphe RAPD– Amplifikationsprodukte von acht Mono-Uredinium-Isolaten wurden auf einem SSCP-Gel getrennt, um Unterschiede zwischen ihnen nachzuweisen. Banden, die bei den acht geprüften Isolaten mögliche polymorphe Loci darstellten, wurden aus dem SSCP-Gel ausgeschnitten und mit PCR reamplifiziert, dann geklont und sequenziert. Für zwei von insgesamt drei sequenzierten DNA-Fragmenten wurde ein Primerpaar entwickelt, um ein in der Grösse für die SSCP-Analyse (<600 bp) geeignetes DNA-Fragment zu amplifizieren. Jedes Primerpaar amplifizierte bei allen acht ursprünglich für ihre Entwicklung verwendeten Isolaten ein PCR-Produkt, und diese wurden anschliessend mit SSCP analysiert. Für beide putativen Loci wurden bei den Isolaten Polymorphismen festgestellt. Die beiden Primerpaare amplifizierten ein PCR-Produkt der erwarteten Grösse bei allen 32 zusätzlich geprüften Mono-Uredinium-Isolaten des Pilzes. Bei den insgesamt 40 geprüften Mono-Uredinium-Isolaten wurden für die Loci A und B 5 bzw. 11 Allele gefunden, und 12 bzw. 34 Isolate erwiesen sich als heterozygot, was durch mehr als zwei Banden auf den SSCP-Gelen angezeigt wurde. Die Spezifität der Primerpaare wurden mit 106 Pilzisolaten aus verschiedenen Taxa geprüft, darunter andere Roste sowie DNA aus gesunden Pappelblättern aus Gewächshauskulturen. DNA-Amplifikationsprodukte der erwarteten Grösse wurden nur erhalten, wenn DNA von Melampsora medusae f. sp. deltoidae präsent war. Die PCR-Amplifikations-Bedingungen mit diesen beiden Primerpaaren wurde so optimiert, dass ein Genotyping direkt bei einzelnen von infizierten Blättern entnommenen Uredinien erfolgen kann und somit eine Pilzkultur zur Charakterisierung von Individuen entfällt. Dies ermöglicht grosse Probenzahlen in Populationsstudien.
1 Introduction
Rusts (Uredinales) are responsible for some of the most damaging diseases of crops and trees, causing important economic losses throughout the world. Poplar leaf rust is considered one of the most important poplar diseases worldwide, causing early defoliation, reduction of growth in height and diameter and, in severe cases, mortality (Toole 1967; Schipper and Dawson 1974; Widin and Schipper 1981; Wang and van der Kamp 1992). Several important questions concerning the epidemiology and population genetics of this rust have yet to be addressed, such as the presence of biotypes within the taxon currently known as Melampsora medusae f. sp. deltoidae (Mmd), the source of primary inoculum, and the importance of the aecial (alternate) host in the epidemics. The use of molecular approaches has the potential to help elucidate some of these questions.
Rust biology and population genetics have been difficult to study in the past because of the biotrophic nature of these fungi. Although landmark studies in well-characterized pathosystems have revealed the contrasting population structures between sexual and asexual cereal rust populations using virulence markers and isozymes (Simons et al. 1979; Roelfs and Groth 1980; Groth and Roelfs 1982; Burdon and Roelfs 1985; Lu and Groth 1988; Linde et al. 1990), the population structure of many rust pathosystems has not yet been characterized.
The advent of restriction fragment length polymorphism (RFLP; Botstein et al. 1980) and random amplified polymorphic DNA (RAPD; Welsh and McClelland 1990; Williams et al. 1990) analyses, allowing the genotyping of large numbers of individuals, has allowed investigators to clarify the population structure of important pathogens (Milgroom and Fry 1997). However, these approaches require a fairly large amount of pure fungal DNA in order to produce DNA profiles. For some rusts, population studies have been conducted by bulking several aecia for RFLP and isozyme analyses (White et al. 1996), by generating axenic cultures (Gitzendanner et al. 1996; Kinloch and Dupper 1996; Kinloch et al. 1998), or by sampling individual aecia or spermogonia directly from cankers (Hamelin et al. 1995; Hamelin 1996; Pappinen et al. 1996; Hamelin et al. 1998). Although these approaches have generated important data, they present problems with regards to sample size and data interpretation. In addition, for some rusts, only the telial host stage is present over large parts of the distribution range and the above-mentioned approaches are not appropriate.
Obtaining large amounts of DNA constitutes a major constraint for population genetics studies of rusts such as Mmd, restricting the possibilities of using markers based on random priming, such as RAPDs, random amplified microsatellites (RAMS; Litt and Luty 1989) or amplified fragment length polymorphisms (AFLPs; Zabeau and Vos 1993; Vos et al. 1995). The only way to cultivate this rust under laboratory conditions is by artificial inoculation of poplar leaves or small poplar plants, which is time consuming, can potentially lead to cross contamination and can impose severe limitations regarding sample size.
An additional constraint in using dominant markers such as RAPDs for Mmd resides in the dikaryotic nature of urediniospores. This type of spore constitutes the only fungal material that can be increased in sufficient amounts to generate enough DNA for fingerprinting analysis. RAPD markers have already been used with dikaryotic spores of another rust, Cronartium ribicola J. C. Fischer (Hamelin et al. 1995; Et-Touil et al. 1999), through statistical corrections in a similar manner as diploids (Lynch and Milligan 1994). However, large sample sizes (up to 2–10 times larger) have to be analysed and several assumptions have to be satisfied, for example concerning null alleles and Hardy-Weinberg equilibrium, which may not apply to all rusts.
Sequence characterized amplified regions (SCARs; Paran and Michelmore 1993) analysed with single-strand conformational polymorphism (SSCP; Orita et al. 1989) can overcome the above-mentioned limitations. It is a powerful method that uses specific primers flanking anonymous polymorphic DNA regions which are amplified by polymerase chain reaction (PCR) and separated by electrophoresis according to their secondary structure to generate codominant markers. These markers, if tested for specificity to the target species, can potentially be used directly from fungal fruiting bodies, such as rust uredinia.
This paper reports the development of two PCR-based DNA markers through SCARs. Two sets of primers were developed that amplify anonymous Mmd DNA fragments of approximately 600 and 500 bp, respectively, and when used with SSCP analysis, allow direct genotyping of rust uredinia without the need for mono-uredinial culturing. This approach should be useful for large-scale population genetic surveys to address the questions raised above.
2 Materials and methods
2.1 Cultivation of Mmd
2.1.1 Plant material
Eastern cottonwood (Populus deltoides) or Jackii hybrid (Populus deltoides × Populus balsamifera) leaves were used as plant material to cultivate the fungus. Small poplar plants were maintained in the greenhouse for Mmd cultivation. They were grown at 22°C with a 16 h photoperiod, fertilized once a week with Plant-Prod® 20-20-20 (Plant Products Co. Ltd., Brampton, ON, Canada) and treated regularly with Pentac® (Sandoz Agro Canada Inc., Mississauga, ON, Canada) against mites.
2.1.2 Fungal material
Mmd isolates originated from either aeciospores or urediniospores. Tamarack (Larix laricina) needles with aecia were collected in Cap-Rouge (Quebec) and placed individually into 1.5 ml microtubes. Infected needles were used the same day they were harvested to inoculate poplar leaves or were kept at 4°C a few days prior to inoculations. Poplar leaves with uredinia were collected at four locations: Cap-Rouge and Villeroy (Quebec), Wickliffe (Kentucky), and Stoneville (Mississippi). Bulks of urediniospores from either one leaf or two to three leaves from the same tree were obtained by gently scraping the leaf surface with a dissecting needle and recovering spores in 1.5 ml microtubes. Spores were used directly for inoculation of poplar leaves or kept in a desiccator under vacuum at 4°C for subsequent inoculations. Urediniospores stored under these conditions remained viable for several months.
2.1.3 Mono-uredinial Mmd propagation
In order to generate rust specific markers, mono-uredinial rust cultures were developed. Poplar leaves located in positions 5–10 (Larson and Isebrands 1971; Sharma et al. 1980) were removed, submerged in deionized water, dried with paper hand towels and cut into 5 cm2 pieces. A piece of paper towel (Wyp All L20, Kimberly-Clark) covering the bottom of a Petri plate was placed in each Petri dish and soaked with deionized water. A piece of leaf was placed upside down on the wet paper. Inoculations were done either with aeciospores or urediniospores. For aeciospore inoculations, a tamarack needle was fixed inside the cover of each Petri dish with white petroleum jelly with the aecium facing down to release aeciospores on the poplar leaf whereas for urediniospore inoculations, urediniospores of a bulk collection were spread on the poplar leaf using a dissecting needle. In either case, the leaf was sprayed with deionized sterile water and the plate was sealed with Parafilm® (American National Can Company, Neenah, WI, USA). Petri plates were incubated at 18°C with a 16 h photoperiod. For the first 2 days of incubation, in the case of aeciospore inoculations, the cover of the Petri plate was tapped a few times with fingers to favour the release of spores. After 2 days, regardless of the type of inoculation, the Parafilm® was removed. Uredinia were fully developed after an additional 8 days of incubation. Cultures were maintained by inoculating new leaves every 10 days. Mono-uredinial isolates were generated by inoculating a new leaf with a single pustule (urediniospores from a single uredinium).
2.1.4 Cultivation on poplar plants
The subculturing of urediniospores in Petri dishes is subject to contamination, especially by molds, because of high humidity and slow degradation of the leaf in the dish. This could be problematic to generate DNA markers with RAPDs because of the universal nature of these markers. In order to avoid contamination of the mono-uredinial isolates and increase the amount of urediniospores for DNA extraction, whole poplar plant inoculations were performed for the last stage of spore increase. Leaves located at positions between 5 and 10 were inoculated by spreading spores under the leaves with the fingertip, wearing a rubber glove. The poplar plant was transferred to a dew chamber at 20°C for 2 days, and then to a growth chamber at 20°C with a 16 h photoperiod until full development of pustules (about 8 days). Usually one round was sufficient to obtain enough spores for DNA extraction. Spores were handled as above. Forty mono-uredinial isolates were generated this way: 18 from Cap-Rouge, 12 from Villeroy, 8 from Stoneville and 2 from Wickliffe.
2.2 DNA extraction
For each mono-uredinial isolate, about 10 mg of urediniospores were needed for DNA extraction. This approximate quantity of spores was transferred in a sterile 1.5 ml microtube and lyophilized. A volume of diatomaceous earth (Sigma Chemical Company, St. Louis, MO, USA) equivalent to that of spores was added to each tube. Spores were ground in 400 μl of Qiagen extraction buffer (100 mM Tris–HCl, pH 9.5, 2% CTAB, 1.4 M NaCl, 1% polyethylene glycol 8000, 20 mM EDTA, pH 8.0, 1%ß-mercaptoethanol). Suspensions were mixed, incubated at 65°C for 1 h, extracted once with 400 μl of phenol : chloroform : isoamyl alcohol (25 : 24 : 1) and centrifuged at 10 000 g for 5 min. The aqueous phase was transferred to a new microtube, precipitated with one volume of cold isopropanol and centrifuged at 10 000 g for 5 min. Resulting pellets were washed with 70% ethanol, air dried overnight and resuspended in 20 μl of TE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, pH 8.0). DNA extracts in TE buffer were stored at −20°C. One : fifty dilutions of DNA extracts in deionized sterile water were prepared as DNA template for RAPDs.
2.3 Identification of DNA polymorphisms by RAPD
DNA fingerprints of the genome of Mmd were generated from eight mono-uredinial isolates representing four locations, using the RAPD method (Welsh and McClelland 1990; Williams et al. 1990). Amplification reactions were performed with two decameric primers (Operon Technologies Inc., Alameda, CA, USA) instead of one, because it has been reported to generate more amplicons of smaller size (Hu et al. 1995) that can be analysed on SSCP gels. RAPD reactions were conducted with primers OPC1 (5′-TTCGAGCCAG-3′) and OPC3 (5′-GGGGGTCTTT-3′). Reactions were carried out in volumes of 12.5 μl consisting of 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 200 μM of each deoxynucleoside triphosphate, 0.2 μM of each decamer primer, 0.5 U of Taq DNA polymerase (Boehringer Mannheim Biochemica, Mannheim, Germany) and 0.5 μl of DNA template, and were done in a MJ Research PTC-60 thermal cycler (MJ Research Inc., Watertown, MA, USA) with conditions as follows: denaturation for 1 min at 94°C, 40 cycles of 94°C for 5 s, 35°C for 10 s, increment of 0.3°C/s until 72°C is reached (+37°C), 72°C for 30 s and increment of 0.2°C/s until 94°C is reached (+22°C). The reactions were terminated with a 4 min extension at 72°C. Amplified band patterns were visualized by electrophoresis in 1.5% (wt./vol.) agarose in TAE buffer (0.04 M Tris–acetate, 1 mM EDTA, pH 8) and staining with ethidium bromide. Polymorphisms were identified for bands under 600 bp, a size appropriate for SSCP analysis and sequencing.
2.4 Fine-level analysis of DNA polymorphisms by SSCP
DNA fingerprints were analysed by SSCP following a protocol adapted from Bagley et al. (1997). For each individual reaction, 5 μl of the RAPD product was mixed with 3 μl of formamide loading buffer (0.05% bromophenol blue, 0.05% xylene cyanol in formamide), denatured at 100°C for 3 min and quickly chilled on ice before gel loading. A volume of 6 μl was loaded on gel and migration was achieved on 0.6× TBE, 0.5× Hydrolink MDETM (FMC® BioProducts, Rockland, ME, USA) non-denaturing polyacrylamide gel (size 260 mm × 125 mm × 0.5 mm) at 2 W for 6 h at 22°C using a Multiphor II system with a MultiTemp II Thermostatic Circulator and a ECPS 3000/150 power supply (Pharmacia, Uppsala, Sweden). SSCP patterns were revealed by silver staining using a Bio-Rad Silver Stain Kit (Bio-Rad Laboratories, Hercules, CA, USA). Bands present in the RAPD profile of some isolates but absent in others were identified.
2.5 Cloning and sequencing of putative polymorphic loci
Gel pieces containing selected bands were recovered with a sterile micropipette tip, resuspended in 20 μl of elution buffer (0.5 M ammonium acetate, 1 mM EDTA, pH 8.0) and heated at 65°C for 15 min. Thirteen bands were extracted representing three putative loci (see Fig. 1). Amplification was performed using 0.5 μl of the suspension as DNA template with conditions described above for RAPDs. Three re-amplified products were selected for cloning on the basis of a well-defined amplification band without secondary bands. Fragments were cloned using the TA Cloning® Kit (Invitrogen®, Carlsbad, CA, USA). Resulting recombinant plasmid DNA was purified with Plasmid Midi Kit (Qiagen, Hilden, Germany), quantified using a Ultrospec 3000 spectrophotometer (Pharmacia, Uppsala, Sweden) and sequenced with an automatic sequencer ABI PRISM® 373A Strech (Applied Biosystems, Foster City, CA, USA) using the BigDye® Terminator v 1.00 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA).
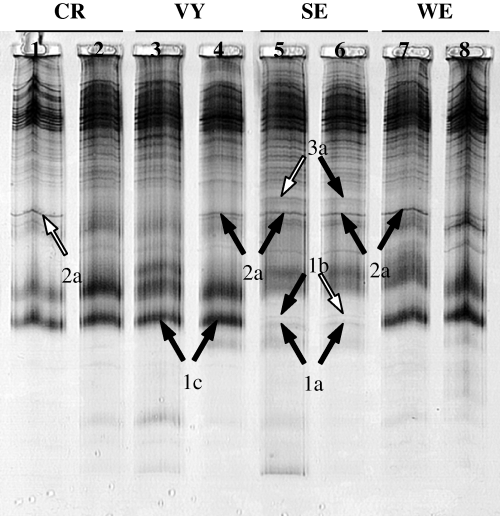
SSCP analysis of RAPD products generated with pair of decamers OPC1 and OPC3 from eight mono-uredinial isolates of Melampsora medusae f. sp. deltoidae representing four locations; Cap-Rouge (CR), Villeroy (VY), Stoneville (SE), and Wickliffe (WE). Selected polymorphic bands, from which DNA was recovered, are indicated with arrows and identified with a two-character code describing the putative loci (numbered 1–3) and the putative alleles for each putative locus (designated by a letter, from a to c, next to the locus number). Fragments that were selected for cloning and sequencing following results of re-amplification from extracted bands are indicated with a white arrow
2.6 Development of primers
Following the analysis of the three DNA fragments sequenced, two fragments (representing two putative loci) were selected for primer development. For each fragment, a primer pair was designed including the decameric priming sites plus the adjacent nucleotides. These primers were then assayed on the eight mono-uredinial isolates that were used to generate them, to eliminate those that did not produce a PCR product on all isolates. One : fifty dilutions of mono-uredinial DNA extracts were used as DNA template. PCRs were performed in 25 μl reaction volumes containing 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM of each dNTP, 1 μM of each primer, 1 U of Taq DNA polymerase and 1 μl of DNA template, using the following program: denaturation for 3 min at 95°C, 30 cycles of 92°C for 30 s, 55°C for 30 s and 72°C for 1 min, and then a final extension at 72°C for 10 min. Primer pairs representing a potential marker were then used on all 40 mono-uredinial isolates to ensure that a PCR product was produced for all isolates. The primer pairs representing markers were tested against 106 fungal isolates from various taxa, including other rusts, and against greenhouse-grown healthy poplar leaves.
2.7 SSCP analysis of markers
Since renaturation and secondary structure depend on DNA sequence, SSCP conditions were individually optimized for each marker to reveal as many different alleles. The parameters used for optimization comprised temperature (4°C, 10°C, 22°C, 32°C) and MDETM concentration (0.5×, 0.8×, 1×). For the marker involving c1c3a2f and c1c3a2r primers (locus A), migration conditions were 0.6× TBE, 0.8× Hydrolink MDETM at 2 W for 18 h at 10°C. For the marker amplified with c1c3a3f and c1c3a3r (locus B), migrations were achieved with 0.6× TBE, 0.8× Hydrolink MDETM at 8 W for 20 h at 4°C.
2.8 DNA amplification from single uredinia
For rust genotyping directly from single uredinia, isolated round-shaped uredinia were selected to avoid confluent uredinia. Urediniospores were removed from uredinia by gently scratching the surface with a sterile dissecting needle. Four cuts were made in the leaf with a sterile scalpel so that a piece of leaf containing the uredinium was removed, placed in a sterile 1.5 ml microtube and used directly for DNA extraction or stored at −80°C until extraction. DNA was extracted as detailed above. Extracts were resuspended in 20 μl of TE buffer (pH 8.0). Because of the small amount of fungal DNA in each extract, 1 μl of extract in TE buffer was added directly to a 12 μl PCR reaction mix composed of 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.4 mM MgCl2, 200 μM of each dNTP, 1 μM of each primer and 0.5 U of Taq DNA polymerase. The amplification program was as described for primer development except that there were five extra cycles to increase yield. Amplification was achieved with the same conditions for both markers.
2.9 Sequence analysis of alleles
All alleles at locus A and seven alleles at locus B were extracted from the Hydrolink MDETM gel, re-amplified using PCR conditions stated in the development of primers section, purified using Qiagen purification columns, and sequenced. The sequences were aligned with BioEdit Sequence Alignment Editor version 5.0.9 (Hall 1999) shareware using Clustal W multiple alignment algorithm (Thompson et al. 1994) and a phylogenetic tree was constructed by using the DNAPARS algorithm in the PHYLIP software package (Felsenstein 1993) to estimate the evolutionary relationship among alleles.
2.10 Segregation analysis of alleles
To obtain isolated basidiospores from a single telium for PCR analysis, a special device was designed: under a biological hood, the inside of a 1 l sterile beaker was first sprayed with deionized sterile water. A 1.5% Bacto agar (Difco Laboratories, Detroit, MI, USA) plate (without its cover) was placed at the bottom of the beaker, while the cover was saved aseptically for further use. Individual telia were recovered from infected poplar leaves collected in the spring, simply by cutting a piece of leaf containing an isolated telium. Using a drop of white petroleum jelly, a piece of leaf with telium was fixed inside the aluminum foil cover that was previously used to autoclave the beaker. The telium was sprayed with deionized sterile water and the cover was put back on the beaker, the telium facing down toward the uncovered Bacto agar plate. This device was put on a rotating agitator at 40 g at room temperature for 10 days. Telia were aseptically recovered in 1.5 ml microtubes and kept at –20°C until DNA extraction and amplification with marker. The Petri plate was taken out of the beaker, the cover was replaced, sealed with Parafilm® and stored a few days at 4°C until observation or directly examined using an inverted microscope Fluovert FS (Leitz, Wetzlar, Germany). Isolated basidiospores were identified on the Petri dish. To amplify a marker from individual basidiospores, 0.6 μl thin-walled PCR tubes containing 25 μl reaction volumes already described for amplification of markers, were prepared prior to picking up basidiospores and kept on ice while recovering basidiospores for amplification. With the help of an inverted microscope Fluovert FS, each isolated basidiospore on a Petri plate was picked up with a piece of Bacto agar using a sterile micropipette tip and released into a different PCR tube with 25 μl reaction volume by pipetting on and off in the reaction mix to evacuate the spore-agar plug from the tip. Each resulting mixture was kept on ice while recovering other basidiospores. Ten to 20 PCR reactions were prepared in each experiment depending on the number of isolated basidiospores available. PCR conditions were exactly those previously described for amplication of markers directly from uredinia.
3 Results
3.1 Identification of DNA polymorphisms using RAPD and SSCP
RAPD profiles obtained from eight mono-uredinial isolates of Mmd with pair of decamers OPC1-OPC3 showed multiple bands covering a wide range of lengths. Fragments amplified ranged from 350 to 2000 bp, and length polymorphisms were identified among isolates. Because some of the fragments showing polymorphisms were below 600 bp, RAPD profiles from this primer pair were chosen for analysis by SSCP.
RAPD fingerprints were separated on a SSCP gel to identify sequence polymorphisms that might not have been visible on agarose gels (Fig. 1). Polymorphic bands (present-absent or length polymorphism) were retained for re-amplification and sequencing. Thirteen bands were removed from profiles generated by OPC1-OPC3 (Fig. 1).
Re-amplification of the 13 DNA fragments from the SSCP gel was successful in most cases (11/13). Three re-amplified products ranging from 300 to 600 bp were selected for cloning and sequencing, based on the presence of a unique band (n = 2) or two bands of different sizes (n = 1).
3.2 Development of markers
Two putative loci (band 2a from Cap-Rouge and 3a from Stoneville; Fig. 1) were selected to develop primer sets for amplification of Mmd markers. For each one of the two selected loci, one primer pair was designed. Each set of primers revealed an amplification product from the eight mono-uredinial isolates used to generate the RAPD profiles. Primer pairs c1c3a2f-c1c3a2r and c1c3a3f-c1c3a3r amplified products of approximately 500 and 600 bp long, respectively, which were further analysed by SSCP. Differences were detected among amplified products of isolates for primer pairs c1c3a2f-c1c3a2r and c1c3a3f-c1c3a3r. These primers yielded an amplification product of the predicted size for all 40 mono-uredinial isolates. The PCR products amplified by primer pairs c1c3a2f-c1c3a2r and c1c3a3f-c1c3a3r were designated locus A and B respectively (Table 1). SSCP analysis revealed 5 alleles at locus A and 11 alleles at locus B for the 40 isolates tested (Fig. 2a, b). When these primers were used in PCR against 80 isolates from other rust species (Chrysomyxa spp., Cronartium spp., Melampsora allii-populina, Melampsora euphorbiae, Melampsora hypericorum, Melampsora larici-populina, Melampsora larici-tremulae, Melampsora pinitorqua, Melampsora rostrupii, Peridermium harknessii, Pucciniastrum spp., Uredinopsis spp.), 26 isolates from various other taxa representing fungal contaminants (Botrytis cinerea, Coelomycetes spp., Coniothyrium spp., Fusarium spp., Gliocladium roseum, Hyphomycetes spp., Pestalotiopsis funerea, Phomopsis sp., Sirococcus conigenus), and DNA extracts from healthy poplar leaves, they did not produce amplicons of the same size as for those obtained for Mmd (data not shown). Of the 40 mono-uredinial isolates tested, 12 were heterozygous at locus A as indicated by the presence of more than two bands on the SSCP gel, while the remaining isolates were homozygous (Fig. 2a). Heterozygosity revealed at locus B was higher, with 34 isolates being heterozygous (Fig. 2b).
| Locus | Primer name | Primer sequence1 | Size of amplified product (bp)2 | Number of alleles3 |
|---|---|---|---|---|
| A | clc3a2f | 5′-GGGGGTCTTTAGGACAAA-3′ | 502 | 5 |
| clc3a2r | 5′-TTCGAGCCAGCATGAAACAC-3′ | |||
| B | clc3a3f | 5′-TTCGAGCCAGAAGTTGTTTC-3′ | 594 | 11 |
| clc3a3r | 5′-TTCGAGCCAGGATCACTT-3′ |
- 1Underlined part of primer sequences corresponds to that of decamers (Operon Technologies Inc., Alameda, CA, USA) which were used to generate the fragment in the first place through a RAPD reaction; OPC1 (5′-TTCGAGCCAG-3′) and OPC3 (5′-GGGGGTCTTT-3′).
- 2Amplified fragments may differ by a few base pairs in size depending on the allele. Indicated fragment sizes are from sequences that were used to design primers.
- 3According to the 40 mono-uredinial isolates tested.
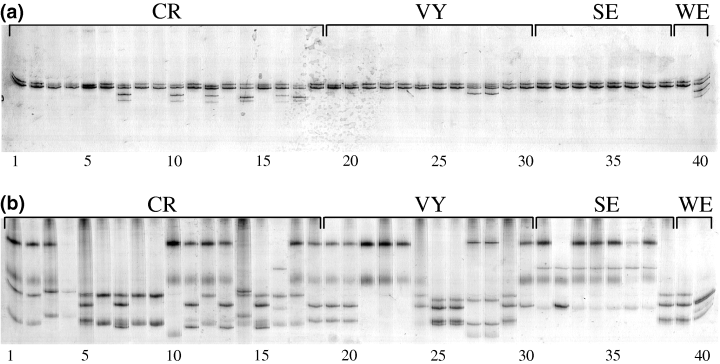
SSCP profiles of all 40 mono-uredinial isolates of Melampsora medusae f. sp. deltoidae generated for this study. (a) Using c1c3a2f and c1c3a2r primers, five alleles were detected; (b) using c1c3a3f and c1c3a3r primers, 11 alleles were detected. With either primer pair, profiles are constituted of two to four bands; homozygotes present two bands whereas heterozygotes present three or four bands. Among the 40 mono-uredinial isolates genotyped, 18 are from Cap-Rouge (CR), 12 from Villeroy (VY), eight from Stoneville (SE), and two from Wickliffe (WE)
3.3 Genotyping from single uredinia
Amplification conditions were optimized to allow genotyping for loci A and B directly from single uredinia, thus alleviating the need to culture the fungus for analysis. Fifteen single uredinia DNA extracts were processed using the optimized conditions (no DNA dilution and five extra PCR cycles). Each extract yielded the expected amplification product with both sets of primers. The testing of the two sets of primers against greenhouse-grown healthy poplar leaves confirmed that secondary bands, previously observed for amplification with primers c1c3a2f-c1c3a2r from single uredinia, resulted from poplar leaf DNA (Fig. 3). These secondary bands migrated at clearly distinct rates in comparison with locus A alleles and did not interfere with genotype interpretation. No PCR product resulted from amplification using c1c3a3f-c1c3a3r with healthy poplar DNA as template (data not shown). Characterization of single uredinia was done by SSCP analysis of both markers. In all cases, only two to four bands were detected on SSCP gels, confirming that a single dikaryotic uredinium was assayed and could be genotyped.
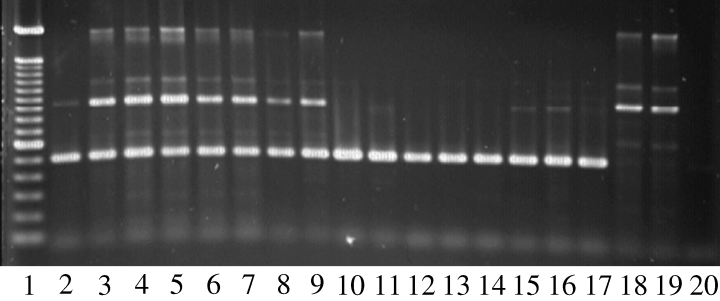
Amplification with c1c3a2f and c1c3a2r primers. Lane 1, 100 bp ladder (Pharmacia); lanes 2–9, amplification from single uredinia of Melampsora medusae f. sp. deltoidae (Mmd) (template DNA = fungal DNA + plant DNA); lanes 10–17, amplification from mono-uredinial isolates of Mmd (template DNA = fungal DNA only); lanes 18 and 19, amplification from healthy poplar leaves (template DNA = plant DNA only); lane 20, negative control (no template DNA)
3.4 Phylogeny of alleles
All five alleles detected at locus A were sequenced, while seven alleles were sequenced at locus B. Allele sequences were deposited into GenBank under the accession numbers AY489127–AY489134 (locus A) and AY490805–AY490813 (locus B). Seven polymorphic sites were observed in the five alleles of locus A (Table 2). Among these polymorphic sites, two revealed so-called allele F to represent in fact two co-migrating alleles, which are indistinguishable from one another under the SSCP conditions used. More polymorphisms were present at locus B with 37 polymorphic sites (Table 3). The relationship among the alleles at locus B was established by phylogenetic analysis (Fig. 4). Two pairs of alleles appear to be closely related. Alleles P and V differ only by a single nucleotide, while alleles F and O share a number of polymorphisms, including a tandem repeat of Ts that is completely conserved. Three alleles at locus A (alleles A, C and F) and two alleles at locus B (alleles B and E) were sequenced from at least two different populations, showing alleles are present in multiple populations, thus supporting the hypothesis that these alleles are not geographic variants.
| Allele1 | Position2 and nucleotides of each polymorphic site | ||||||
|---|---|---|---|---|---|---|---|
| 36 | 274 | 331 | 372 | 377 | 408 | 470 | |
| A | T | T | C | C | A | G | C |
| B | T | C | G | T | A | G | C |
| C | G | T | C | C | A | G | C |
| D | T | C | C | C | A | G | C |
| F #1 | T | T | C | C | A | G | G |
| F #2 | T | T | C | C | G | A | C |
- 1The allelic pattern identified as allele F on SSCP gel happened to represent co-migration of two distinct alleles, F #1 and F #2.
- 2Positions of polymorphic sites are calculated from the first nucleotide at the 5′-end of c1c3a2f primer, considered position 1 in the alignment of all alleles. Alignment ends at position 502, which corresponds to 5′-end of reverse (c1c3a2r) primer. Allele sequences from this alignment are available from GenBank (AY489127–AY489134).
| Allele | Position1 and nucleotides2 of each polymorphic site | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 51 | 52 | 67 | 79 | 84 | 92 | 93 | 102 | 103 | 112 | 114 | 131 | 161 | 186 | 195 | 196 | 197 | 198 | 199 | 200 | 201 | 202 | 205 | 225 | 230 | 249 | 276 | 304 | 306 | 328 | 369 | 370 | 382 | 415 | 419 | 439 | 490 | |
| B | C | A | G | A | C | C | A | A | T | A | C | G | T | C | – | – | – | – | – | – | – | – | T | G | T | A | C | A | G | G | T | G | C | C | C | T | C |
| D | C | A | G | A | C | C | G | A | G | A | T | A | C | C | – | – | – | – | – | – | T | T | T | G | T | A | C | T | A | A | C | – | C | T | C | C | C |
| E | C | A | G | A | C | C | G | A | G | A | T | A | C | C | – | – | – | – | – | – | – | – | T | G | T | A | C | T | A | A | C | – | C | T | C | C | C |
| F | T | G | C | G | T | G | A | C | G | A | T | G | C | C | T | T | T | T | A | T | T | T | A | G | G | A | T | A | A | A | A | – | C | C | T | C | C |
| O | T | G | C | A | T | G | A | A | G | C | T | G | C | A | T | T | T | T | A | T | T | T | A | A | G | G | T | A | A | A | A | – | T | C | T | C | C |
| P | C | A | G | A | C | C | G | A | G | A | T | A | C | C | – | – | – | – | – | – | – | T | T | G | T | A | C | T | A | A | C | – | C | T | C | C | T |
| V | C | A | G | A | C | C | G | A | G | A | T | A | C | C | – | – | – | – | – | – | – | T | T | G | T | A | C | T | A | A | C | – | C | T | C | C | C |
- 1Positions of polymorphic sites are calculated from the first nucleotide at the 5′-end of c1c3a3f primer, considered position 1 in the alignment of all seven alleles. Alignment ends at position 603, which corresponds to 5′-end of reverse (c1c3a3r) primer. Allele sequences from this alignment are available from GenBank (AY490805–Y490813).
- 2Dashes refer to the absence of nucleotide as a consequence of alignment.
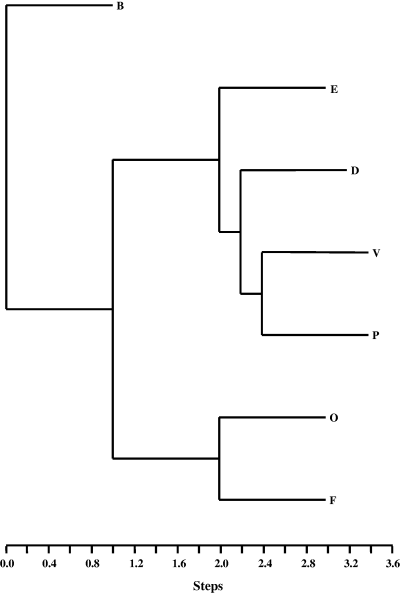
Phylogenetic relationship of seven alleles from locus B generated by the method of parsimony using DNAPARS algorithm from PHYLIP software package. Two alleles were sequenced from samples originating from two different populations: allele B from Canada, Quebec, Villeroy (AY490805) and USA, Mississippi, Stoneville (AY490806), and allele E also from Canada, Quebec, Villeroy (AY490808) and USA, Mississippi, Stoneville (AY490809)
3.5 Segregation of alleles
Only two experiments (one per locus) yielded enough amplification products to reveal each allele of a heterozygote telium. Telium F6TE3 is heterozygous for alleles A and D at locus A. The results from the basidiospore amplification revealed that there was segregation for the two alleles at this locus. Seven individual basidiospores yielded a PCR product at that locus. Three basidiospores revealed the pattern of allele A, while four basidiospores possessed allele D (Fig. 5a). Although sample size was small due to the difficulty in obtaining and amplifying single basiodiospores from heterozygous telia, the results are not significantly different from the expected 1 : 1 segregation ratio (p > 0.05). At locus B, telium F2T20 is heterozygous for alleles B and C. Here again, the basidiospore amplification revealed that there was segregation for the two alleles at this locus. Four individual basidiospores yielded a PCR product at that locus. One basidiospore revealed the pattern of allele B, while three basidiospores possessed allele C (Fig. 5b).
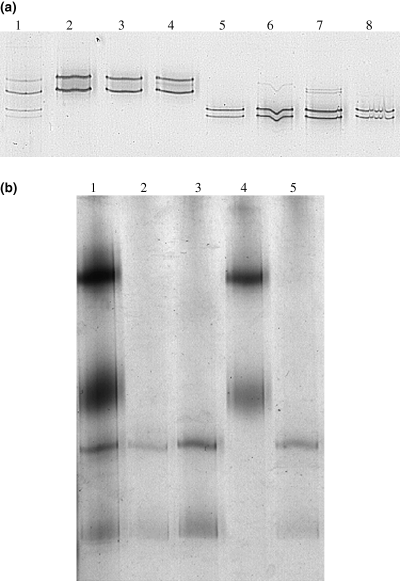
Segregation analysis of alleles at locus A and B. (a) SSCP profiles of marker at locus A for telium F6TE3 and its progeny basidiospores. Lane 1, heterozygote telium with alleles A and D; lanes 2–4, 3 individual basidiospores with allele A; lanes 5–8, 4 individual basidiospores with allele D. (b) SSCP profiles of marker at locus B for telium F2T20 and its progeny basidiospores. Lane 1, heterozygote telium with alleles B and C; lanes 2, 3 and 5, 3 individual basidiospores with allele C; lane 4, 1 individual basidiospore with allele B
4 Discussion
There are two main constraints to population genetic studies of rusts. The first one relates to the biotrophic nature of these fungi. To obtain genetic data with isozymes, RFLPs or RAPDs, mono-uredinial cultures have to be produced on host leaves, a long, tedious and non-sterile process which may lead to cross contamination and undesirable selection processes during subculturing. Although axenic cultures have been successfully produced for some rusts (Williams et al. 1967; Quick and Cross 1971; Raymundo and Young 1974; Hu and Amerson 1991; Kinloch and Dupper 1996), they involve the haploid stage of the rust which can be difficult to obtain. In addition, axenic cultures would be difficult to generate for large-scale population studies.
The second constraint relates to the fact that the most abundant and most widely used spore stages for rust population studies are the dikaryotic urediniospores or aeciospores. This causes a problem when dominant markers such as RAPDs are used. Although haploid spore stages (spermatia) have been used to study white pine blister rust (Hamelin 1996; Hamelin et al. 1998) and fusiform rust (Doudrick et al. 1993; Hamelin et al. 1994), spermatia are very cryptic or absent in other rusts. Statistical corrections for diploid (or dikaryotic) material (Lynch and Milligan 1994) have also been used for analysis of RAPDs for white pine blister rust (Hamelin et al. 1995). However this approach does not allow the establishment of haplotypic identity because of marker dominance, nor does it allow assessment of observed heterozygosity and tests of Hardy-Weinberg equilibrium.
The method we describe here addresses both of these constraints by assigning codominant genotypes directly from uredinia on infected leaves. We have some evidence that the alleles identified belong to single loci. Although segregation studies were limited by the difficulty in obtaining segregating populations, we performed SSCP analyses on single basidiospores released from a unique telial column in a Petri dish. In all instances, a single allele was present in each basidiospore, a result that would be unexpected if the alleles belonged to different loci (Fig. 5). Additionally, sequence analysis indicates a high level of homology (>90%) among alleles at each of these two loci and allele phylogeny did not indicate the presence of highly divergent clades which could have indicated the presence of multiple loci. Finally, DNA fragments amplified with either set of primers generated SSCP patterns composed of two (homozygotes), three or four (heterozygotes) bands for each dikaryotic mono-uredinial isolate (Fig. 2a, b). Given the large number of alleles, the presence of no more than two alleles per individual would be highly unlikely if the alleles were from paralogous loci. All the evidence presented here is consistent with an interpretation of the alleles at these loci as codominant.
The approach we report here allowed us to find two markers for Mmd. Although a necessary prerequisite for the process of developing markers was the cultivation of mono-uredinial isolates of the fungus to obtain enough high-quality DNA for screening the genome, the PCR-SSCP marker assay we developed was used directly with uredinia without the need to cultivate the fungus. Genotyping directly from the uredinia should be an extremely powerful tool, allowing population studies on very large numbers of samples at various hierarchical levels (e.g. within leaves, among leaves within trees, among trees, etc.). This could be a powerful approach to conducting association studies directly from the phenotypes of the host-pathogen interaction.
The use of two decamers in a RAPD reaction as compared to one has been reported to generate smaller fragments (Hu et al. 1995). Such an approach thereby increases the probability of detecting polymorphisms for small fragments that are suitable for SSCP analysis and DNA sequencing, and reduces the number of primers to be screened. Separation of RAPD profiles by SSCP enhanced the discovery of polymorphisms and also contributed to the isolation of DNA fragments for cloning and sequencing. This could be important for other fungi where levels of polymorphisms are low.
We used RAPD and SSCP to identify polymorphisms and target fragments. Such a complementary set of analyses is not essential for that purpose but seemed preferable to us. RAPD analysis or variations of the technique based on single repeat, random amplified microsatellites (RAMS) can be used alone for detecting and targeting polymorphic fragments (Groppe et al. 1995; Dusabenyagasani et al. 1998). However, migration of RAPD products by standard electrophoresis on agarose or polyacrylamide gel shows less sensitivity for detecting polymorphisms than migration in SSCP conditions (Hayashi 1991; McClelland et al. 1994). Also, extracting bands is often complicated in agarose gels by the presence of co-migrating DNA fragments. SSCP analysis of RAPD products may reveal polymorphisms undetected by standard electrophoresis methods, as migration of single-stranded DNA under SSCP conditions allows detection of single mutations (Orita et al. 1989). Combination of RAPD and SSCP analyses for marker generation avoids extensive screening for polymorphisms in the case of organisms showing few polymorphic bands in RAPD profiles.
The new markers reported here will be useful to answer molecular epidemiology questions such as the source of inoculum of poplar leaf rust epidemics and the importance of host alternating and asexual reproduction in the life cycle of this rust. The markers will generate population-based sequence data of a codominant nature and will allow the calculation of Hardy-Weinberg equilibrium, linkage disequilibrium and other population genetics parameters. Moreover, the evolutionary relationship among the alleles, established by generating allele phylogenies, provides useful information on the evolutionary relationship among individuals. This will provide tools to test hypotheses about the type of spores involved in rust dissemination (sexual vs. asexual) and will help in tracking continental spore movement.
Acknowledgements
The authors would like to thank F. Caron and P. Périnet from the ministère des Ressources naturelles du Québec, Villeroy, Québec, T. Robison and R. Rousseau from Westváco, Wickliffe, Kentucky, D. Wilson from USDA Forest Service, Southern Research Station, Centre for Bottomland Hardwoods Research, Stoneville, Mississippi, for providing samples. Dr. P. Frey from INRA Pathologie forestière, Champenoux, France, tested the markers on different Melampsora species. We also thank N. Lecours for providing laboratory assistance and M. Dusabenyagasani for reviewing the manuscript. This study was supported by USDA Competitive Grant 94-37303-0735.




