Expression of the RNA recognition motif protein RBP10 promotes a bloodstream-form transcript pattern in Trypanosoma brucei
Summary
When Trypanosoma brucei differentiates from the bloodstream form to the procyclic form, there are decreases in the levels of many mRNAs encoding proteins required for the glycolytic pathway, and the mRNA encoding the RNA recognition motif protein RBP10 decreases in parallel. We show that RBP10 is a cytoplasmic protein that is specific to bloodstream-form trypanosomes, where it is essential. Depletion of RBP10 caused decreases in many bloodstream-form-specific mRNAs, with increases in mRNAs associated with the early stages of differentiation. The changes were similar to, but more extensive than, those caused by glucose deprivation. Conversely, forced RBP10 expression in procyclics induced a switch towards bloodstream-form mRNA expression patterns, with concomitant growth inhibition. Forced expression of RBP10 prevented differentiation of bloodstream forms in response to cis-aconitate, but did not prevent expression of key differentiation markers in response to glucose deprivation. RBP10 was not associated with heavy polysomes, showed no detectable in vivo binding to RNA, and was not stably associated with other proteins. Tethering of RBP10 to a reporter mRNA inhibited translation, and halved the abundance of the bound mRNA. We suggest that RBP10 may prevent the expression of regulatory proteins that are specific to the procyclic form.
Introduction
Trypanosoma brucei is an extracellular parasite that belongs to the order of Kinetoplastida. It causes African sleeping sickness in humans, and debilitating diseases of domestic animals, thereby constituting a burden to public health and economies of many countries in sub-Saharan Africa.
Trypanosomes are transmitted between mammalian hosts by Tsetse flies. In order to adapt to the environments of the different hosts the parasites have to control their gene expression tightly. Up to 25% of all transcripts are regulated during transformation of mammalian bloodstream forms to the procyclic forms that grow in the Tsetse midgut (Jensen et al., 2009; Kabani et al., 2009; Queiroz et al., 2009; Siegel et al., 2010); the proportion detected varies depending on the technique used for measurement, and the thresholds applied. Some of the gene regulation that is seen during differentiation is required in order for the parasites to adapt to new energy sources. In the human host, the bloodstream form of the parasite derives its ATP from glucose catabolism by glycolysis. The glucose is taken up into the cell mainly by the glucose transporter THT1 (Bringaud and Baltz, 1993) and utilized in the glycosomes, which are microbodies containing most of the glycolytic enzymes. Within the Tsetse fly, proline is thought to be an important energy source, although the evidence for this is extraordinarily tenuous (Bursell et al., 1973; Balogun, 1974). Procyclic trypanosomes certainly use proline when no glucose is present (Lamour et al., 2005), and indeed grow better using proline alone than when supplied mainly with glucose (Ebikeme et al., 2008). In vitro, the differentiation from bloodstream forms to procyclic forms can by triggered by the addition of cis-aconitate to the medium (Overath et al., 1986) and a shift to 27°C; differentiation is facilitated by cis-aconitate transporters (Dean et al., 2009), and involves a signalling pathway that includes protein phosphatases (Szoor et al., 2006; 2010). Interestingly, the removal of glucose alone is also sufficient for the cells to start differentiation (Milne et al., 1998) and a similar effect is seen after inhibition of the glucose transporter using phloretin (Haanstra et al., 2011). During differentiation, the trypanosomes' surface coat of variant surface glycoprotein is replaced by EP and GPEET procyclins.
Despite the need for extensive gene regulation, transcription by RNA polymerase II is not regulated in trypanosomes. Instead, control over gene expression relies largely on changes in mRNA stability and translation (Fernández-Moya and Estévez, 2010). The stabilities of trypanosome mRNAs – as in other eukaryotes studied so far – appears to be determined largely by sequences in their 3′ untranslated regions (3′ UTR). It is assumed that the regulatory 3′ UTR sequences are recognized by RNA binding proteins, which in turn communicate with the degradative and translational machineries (Fernández-Moya and Estévez, 2010; Kramer and Carrington, 2011). Some examples of this are already known. Experiments to investigate regulatory proteins have so far focussed on proteins with known RNA binding domains. For example, the pumilio-domain protein PUF9 stabilizes its target mRNAs (Archer et al., 2009), while UBP1, a protein containing an RNA recognition motif (RRM) has been shown to have either stabilizing or destabilizing effects (D'Orso and Frasch, 2001; Hartmann et al., 2007). ZC3H20 has been shown to bind to, and stabilize, a small number of mRNAs in procyclic forms (Ling et al., 2011). On the other hand, ZFP3, a protein with a CCCH zinc finger domain, has been shown to play a role in translation (Walrad et al., 2009).
The RRM is a sequence of around 90 amino acids, which can bind either to RNA or other proteins (Samuels et al., 1998; Maris et al., 2005). In a previous survey of trypanosome proteins with RRM domains, we identified RBP10 as being essential to the growth of bloodstream-form trypanosomes (Wurst et al., 2009). We also showed that RBP10 mRNA is co-regulated with several mRNAs encoding enzymes of glucose metabolism (Queiroz et al., 2009). In this study, we investigate the function of RBP10 in more detail. We find that the presence of RBP10 is sufficient to induce the expression of mRNAs involved in bloodstream-form energy metabolism; however, the evidence implies that RBP10 may act indirectly, perhaps through inhibition of the translation of other regulators.
Results
RBP10 is a bloodstream-form-specific cytoplasmic protein
RBP10 is a protein of 32 kDa with a single RRM domain (amino acids 49–121). RBP10 RNAi was previously found to be lethal in bloodstream-form trypanosomes (Wurst et al., 2009). In those experiments, we had used an inducible RNAi vector with two opposing T7 promoters. We repeated the experiments by inducibly expressing an RNA stem-loop. In bloodstream-form trypanosomes, the knock-down of RBP10 was lethal after 4 days (Fig. 1A), while no effect on proliferation could be seen in the procyclic form (data not shown). Attempts to knock out the RBP10 gene in procyclic forms were not successful: this may indicate a residual function for RBP10 in that form, or could have been a technical failure.
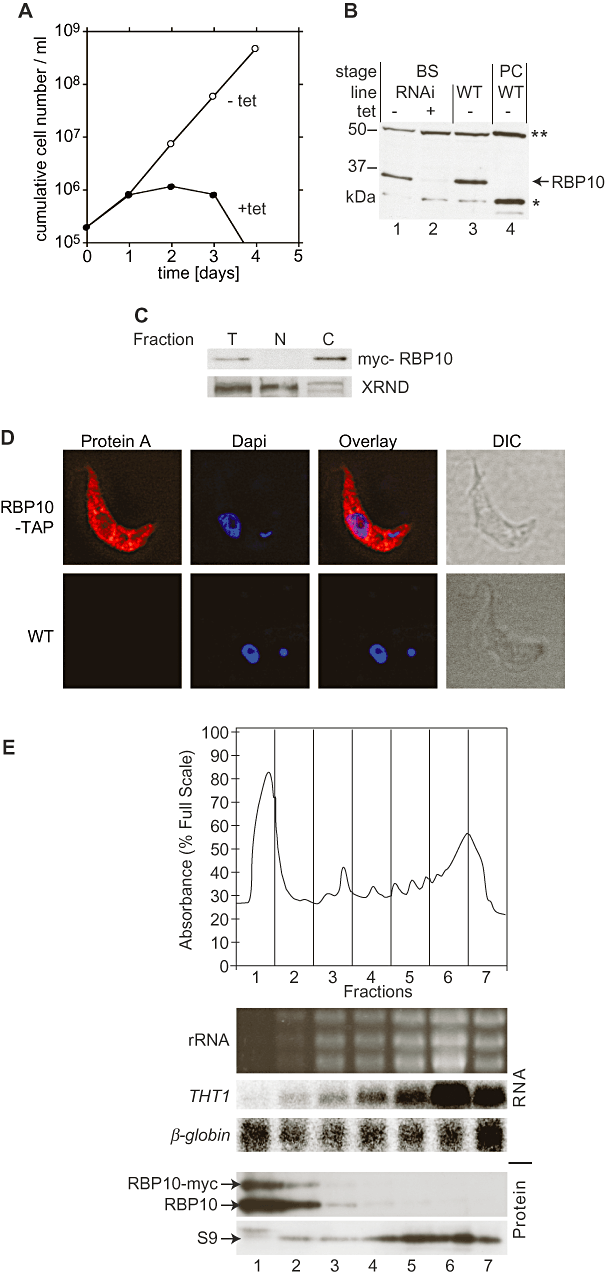
RBP10 is an essential cytoplasmic protein in bloodstream forms. A. Bloodstream-form trypanosomes with tetracycline-inducible RNAi targeting RBP10 were gown in the presence (closed circles) or absence (open circles) of 100 ng ml−1 tetracycline, with dilution as required. A cumulative growth curve is shown. B. Western blot with extracts from bloodstream-form (BS, lanes 1–3) or procyclic-form (PC, lane 4) trypanosomes. The RNAi cell line (lanes 1and 2) was grown with (lane 2) or without (lane 1) tetracycline. Bands corresponding to RBP10, and two cross-reacting proteins (**) and (*) are indicated. C. RBP10-myc is mainly in the cytoplasm. A bloodstream-form cell extract (T) was separated into nuclear (N) and cytoplasmic (C) fractions. The nuclear protein XRND serves as a control. D. RBP10-TAP was expressed in bloodstream-form trypanosomes, and detected by immunoflourescence using anti-IgG antibody. The nucleus and kinetoplast were detected using DAPI, and the cells by differential interference contrast microscopy (DIC). E. Distribution of RBP10 on a sucrose gradient. Polysomes were separated as described in Djikeng et al. (2003). RNA and protein were prepared from equal amounts of each fraction. Before the RNA preparation, a small amount of human beta-globin RNA was added to each fraction as an internal control for RNA isolation efficiency. The upper panel is an OD280 trace. The lower panels are a Northern blot (original ethidium stain showing rRNA, and blot probed for THT1 and human beta-globin control) and a Western blot probed with antisera to RBP10 and to ribosomal protein S9 (C. Helbig, ZMBH, unpublished).
An antibody against full-length recombinant RBP10 recognized a protein of the expected size on Western blots, but also cross-reacted with two other proteins, the larger of which (Fig. 1B, asterisks) appeared to be in the mitochondrion as judged by digitonin fractionation and immunofluorescence (not shown). Various attempts to remove this cross-reactivity failed. By Western blotting, we could see that RBP10 was well expressed in bloodstream forms (Fig. 1B, lanes 1 and 3) but was absent in procyclic-form parasites (Fig. 1B, lane 4). RNAi against RBP10 resulted in strong depletion of the presumed RBP10 protein band (Fig. 1B, lane 2), confirming the specificity of the antibody.
To assess the location of RBP10, we subjected cells to fractionation (Fig. 1C) or immunoflourescence (Fig. 1D). For the fractionation, the staining of XRND, a nuclear protein (Li et al., 2006), served as a control. Additionally, immunofluorescence with an overexpressed RBP10-TAP was done. Both techniques showed RBP10 to be in the cytoplasm; a digitonin fractionation (not shown) confirmed that the protein was in the cytosol. To find out whether RBP10 was associated with polysomes, we subjected extracts to sucrose gradient fractionation (Fig. 1E). The polysomal peak was detected using antibodies to ribosomal protein S9, and hybridization with the mRNA encoding THT1, a bloodstream-form-specific glucose transporter; the fractions were spiked with beta-globin mRNA as a control for mRNA yield. RBP10 was concentrated in the top, soluble protein fraction, trailing down as far as the monosomes (Fig. 1E, fraction 3). No RBP10 was detected in the polysomal peak and there was almost no overlap between RBP10 and the THT1 mRNA.
Effects of RBP10 RNAi on the transcriptome
To examine the global effect of RBP10 depletion in bloodstream-form trypanosomes we performed a microarray analysis, comparing RNA from wild-type parasites (WT, no RNAi plasmid) to bloodstream-form RBP10 RNAi cells 24 h after induction of RNAi. Six slides, including three biological replicates and dye-swap, were analysed. We considered all mRNAs with a P-value ≤ 0.05 and a minimum of 1.5-fold change to be significantly regulated, and re-tested some by Northern blotting (2, 3 and Table S1). Five hundred and ninety-five RNAs were changed by RBP10 RNAi: 275 increased and 320 decreased. Striking changes in mRNA level were observed for genes involved in sugar metabolism and transport (2, 3 and Table S1). Seven of the ten most downregulated mRNAs fell into this category (Table S1), and of the core enzymes of glucose and glycerol metabolism, only the mRNAs encoding fructose bisphosphate aldolase and phosphofructokinase were not at least twofold reduced. Twenty-one mRNAs encoding glycosomal proteins (including glycosomal membrane proteins and enzymes of purine salvage) or proteins involved in glucose metabolism, including glycosomal phosphoglycerate kinase (PGKC) (Fig. 3C) and the major bloodstream-form glucose transporter, THT1, were decreased, as was mRNA encoding the VSG (Fig. 3A). Also broadly affected were genes encoding proteins of flagellum and the cytoskeleton. Meanwhile, an increase in the mRNA encoding EP procyclin was detected (Fig. 3B and Table S1).
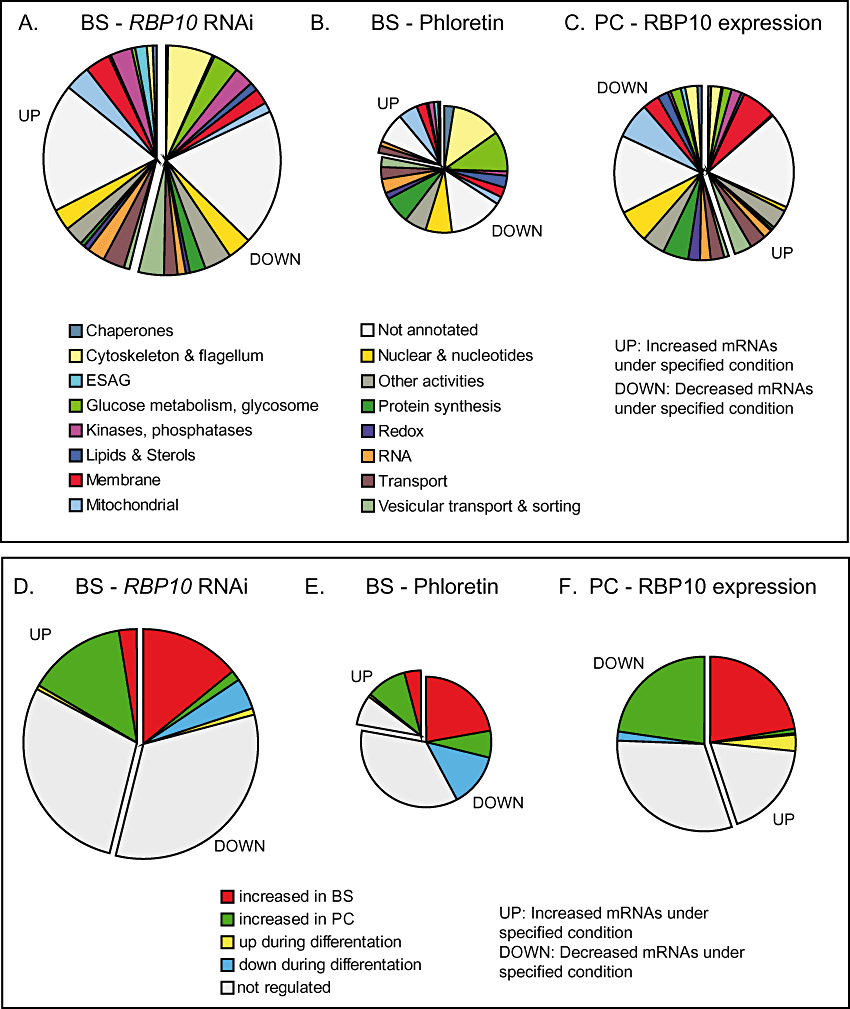
Effects of alterations in RBP10 expression on the transcriptome. A. Effect of RBP10 RNAi in bloodstream forms. Different classes of gene function are shown in different colours. The areas of the segments are to scale. B. RNAs affected by phloretin treatment (Haanstra et al., 2011) on the transcriptome of bloodstream forms; the area of the circle is to scale relative to (A). C. Effect of induced RBP10-myc expression in procyclic forms. Details is in (A) and (B). D–F. As (A–C), but with genes classified according to developmental regulation pattern.

Effect of RBP10 depletion on expression of selected genes. RNA was detected by Northern blotting. All cells were bloodstream-forms unless indicated otherwise (panel B). Each panel shows a separate experiment. Quantification relative to the 7SL control is beneath each relevant lane.
These effects on mRNA levels were all very reminiscent of the early stages of trypanosome differentiation, but could also have been due simply to growth inhibition. To find out if there really was a correlation between the effects of RBP10 depletion and differentiation, we compared the transcriptome results with those of our previous microarray analysis of differentiation (Queiroz et al., 2009) (We restricted ourselves to this dataset because the experimental and statistical methods used were identical to those in the current study, which made quantitative comparisons meaningful). Decreases in glucose uptake, for example after phloretin treatment, have also been shown to decrease expression of glycolytic enzyme mRNAs (Haanstra et al., 2011). Because we had observed a decrease in THT1 mRNA, so expected a reduction in glucose transport, we also compared our results for RBP10 with those seen after phloretin treatment (Haanstra et al., 2011). In our discussion, we will use the term ‘procyclic-form-specific’ to mean simply that the mRNA is more abundant in procyclic forms than in bloodstream forms, and vice versa for ‘bloodstream-form-specific’. Of the 320 mRNAs decreased by RBP10 RNAi, 85 were also bloodstream-form-specific, while only eight were procyclic-form-specific (Fig. 2D). Phloretin treatment did not have such strong effects: 153 mRNAs were decreased including 44 bloodstream-form- and 13 procyclic-form-specific ones (Fig. 2B and E). Seventy-one mRNAs were decreased after both RBP10 RNAi and phloretin. These included 18 mRNAs encoding for proteins of glucose metabolism and/or the glycosome and 16 mRNAs encoding cytoskeletal or flagellar proteins (Table S1). Cytoskeletal and flagellar protein mRNAs are co-ordinately downregulated during growth arrest in the early stages of differentiation (Queiroz et al., 2009). Decreases in these and other proteins that are normally required for growth could either be a consequence, or a cause of the growth arrest seen upon RBP10 depletion.
Two hundred and seventy-five mRNAs increased after RBP10 RNAi. Of these, 84 were procyclic-form-specific and 15 were bloodstream-form-specific (Fig. 2D). Among the upregulated mRNAs were mitochondrial and trans-membrane proteins (Fig. 2A), and also the transporter PAD1, which is upregulated in the stumpy form of the parasite (Dean et al., 2009). The mRNAs encoding two potential RNA binding proteins that are required only in procyclic forms – (Alsford et al., 2011) – ZC3H20 and DRBD6A – also increased. Some targets of ZC3H20 – MCP12 (Tb927.10.12840), Tb927.5.4020 (hypothetical) and a GRESAG4 mRNA – were increased after RBP10 RNAi, but others were not. Thus the effects of changing RBP10 clearly extend beyond those caused by upregulation of ZC3H20.
Fewer (44) mRNAs were increased after phloretin treatment. Among them 20 were procyclic-form- and eight bloodstream-form-specific (Fig. 2E). Overall, 91 mRNAs were affected by both RBP10 RNAi and phloretin; the correlation coefficient within this subset is 0.9. Thus this subset of mRNAs is affected similarly under both conditions. The RNA encoding the regulatory phosphatase PIP39 (Szoor et al., 2010), which increases during differentiation, also increased approximately 1.5-fold after both phloretin treatment and RBP10 RNAi: although the P-values for both PIP39 genes were below the cut-off for phloretin treatment, one of them (Tb09.160.4460) was significantly increased after RBP10 RNAi. The gene encoding another regulatory phosphatase, PTP1 (Tb927.10.6690) (Szoor et al., 2006), was not represented on our microarray.
Expression of RBP10 induces bloodstream-form-specific mRNAs in procyclic forms
RBP10 was not detectable in procyclic-form trypanosomes (Fig. 4C, lane 2). Indeed, we found that anomalous expression of myc-tagged RBP10 strongly inhibited growth (Fig. 4A). Because the transgenic mRNA was present as two species, with shorter 3′ UTRs than the RBP10 mRNA, the transgenic mRNAs could be distinguished readily from the endogenous one (Fig. 4B, lane 3). Similarly, RBP10-myc migrates slower than the untagged protein (Fig. 4C, compare lanes 3 and 5 with lane 1). We could therefore see that ectopic expression of RBP10-myc caused an increase in endogenous RBP10 mRNA (Fig. 4B, top panel, lane 3) and protein (Fig. 4C, lanes 3 and 5; the lower band is once again a cross-reacting protein).
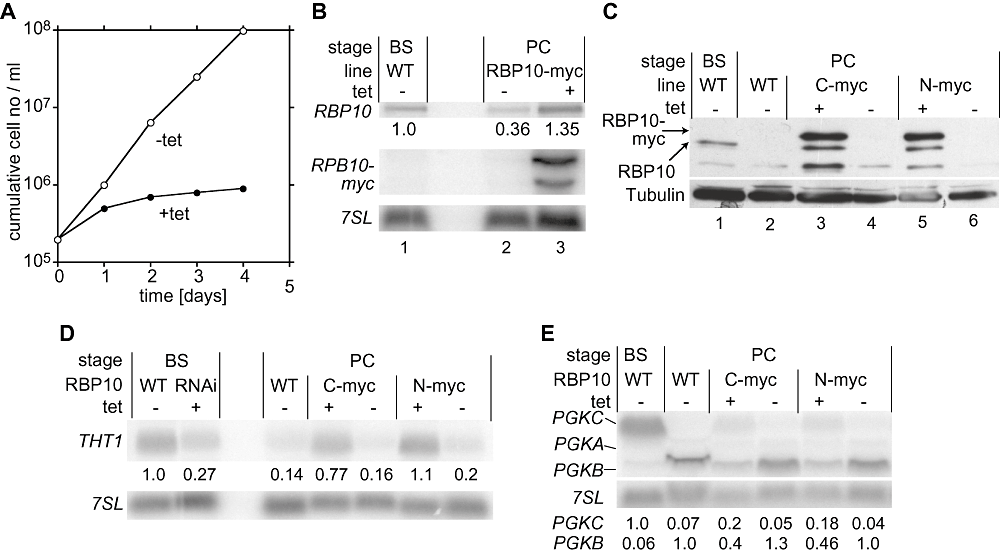
Effects of RBP10 expression in procyclic trypanosomes. A. Effect of expression of RBP10-myc on procyclic trypanosome growth. Details as in Fig. 1. B. Northern blot showing the levels of mRNAs encoding RBP10-myc (1.5 kb and 1.8 kb) and RBP10 (9 kb). Expression of exogenous RBP10-myc was induced 24 h before cells were harvested using 100 ng ml−1 tetracycline. 7SL serves as loading control. C. Forced expression of tagged RBP10 induces expression of endogenous RBP10. Procyclic trypanosomes with that inducibly express C-terminally myc-tagged RBP10 (C-myc) or N-terminally myc-tagged RBP10 (N-myc) (both migrating at 36 kDa) were incubated with or without tetracycline. RBP10 was detected by Western blotting. RBP10 migrates at 33 kDa. Tubulin serves as a control. D. Effect of forced expression of RBP10 on THT1 mRNA levels. E. Effect of forced expression of RBP10 on PGK mRNA levels.
We next investigated the effect of RBP10 expression on procyclic gene expression. The BS-specific THT1 mRNA decreased to about 30% by RBP10 RNAi in bloodstream-form trypanosomes (Fig. 4D). In procyclic forms, where THT1 is normally hardly detectable, expression of RBP10 raised the amount of THT1 to the abundance of bloodstream-form WT (Fig. 4D). A similar effect could be seen for the developmentally regulated PGKB and PGKC mRNAs (Fig. 4E). Usually PGKC mRNA is much more abundant than PGKB in bloodstream forms, while the converse situation pertains in procyclic forms. The expression of RBP10 in procyclic forms led to an increase of PGKC and a decrease in the PGKB. The effect of RBP10 expression was, however, much stronger on THT1 than on the PGK mRNAs.
Microarray results comparing procyclic-form WT RNA with RNA of procyclic forms expressing RBP10-myc yielded results that were in many ways a mirror-image of those obtained by RBP10 RNAi (Table S1). Three hundred and forty-six mRNAs were changed (Fig. 2C) and once again there was a strong correlation with developmental regulation (Fig. 2F). Among the 156 increased mRNAs, 78 were bloodstream-form-specific, and only three were procyclic-form-specific (Fig. 2F). We not only confirmed the increase of the glucose transporter THT1, but also increases in mRNAs encoding for five proteins involved in glucose metabolism: glucose 6-phosphate isomerase, glycerol kinase, ATP-dependent phosphofructokinase, fructose-bisphosphate aldolase and aquaglyceroporin. Additional mRNAs encoding glycosomal proteins were also increased but had P-values higher than the cut-off. Many of these mRNAs show very similar regulation to the RBP10 mRNA during differentiation, suggesting that they all are within the same post-transcriptional regulon (Queiroz et al., 2009). The appearance of RBP10 protein upon RBP10-myc expression in procyclics (Fig. 4C) therefore reflects the overall pattern of induced expression of bloodstream-form-specific mRNAs in this life cycle stage. The results also indicated that both forms of myc-tagged RBP10 were functional.
Several mRNAs for transporters and other trans-membrane proteins were also increased by forced RBP10 expression (Fig. 2C and Table S1), indicating that the change in glucose transport was not the only effect on metabolite uptake. Of the 190 mRNAs that decreased during forced RBP10 expression, 79 were normally procyclic-form-specific, while none was BS-specific (Fig. 2F). A broad decrease of mRNAs encoding mitochondrial components (Fig. 2C) was the inverse of the increase seen during bloodstream-form RNAi. Seven of the decreased mRNAs encoded glycosomal or glycolytic proteins, but three of them were involved in procyclic-form-specific pathways. Another, HK2, is a hexokinase-like protein that may lack enzymatic activity; procyclic-form cells lacking HK2 actually show increased hexokinase activity (Morris et al., 2006). A decrease in mRNAs involved in protein synthesis was most likely a secondary effect of the growth inhibition.
Given these effects on mRNAs involved in energy metabolism, we wondered if the growth defect in the RBP10-expressing procyclics was caused by a failure to generate energy from proline, which was the principal energy source in the culture medium. We therefore attempted to rescue the growth defect by the addition of 4.6 g l−1 glucose; but this did not work (data not shown).
Inhibition of differentiation
Expression of RBP10 correlated with expression of bloodstream-form-specific mRNAs, and RBP10 is bloodstream-form-specific too. We therefore asked whether bloodstream-form trypanosomes that were forced to express RBP10 would be able to differentiate into procyclic forms. We therefore transfected differentiation-competent trypanosomes with an RBP10-myc expression plasmid. Expression was induced with tetracycline for 24 h, then we induced differentiation by adding cis-aconitate and decreasing the temperature to 27°C. As we knew that RBP10 expression would kill procyclic forms, the medium was changed to procyclic medium without tetracycline or cis-aconitate 24 h later. In WT cells, the amount of RBP10 decreased after 24 h in differentiation conditions, and it was undetectable by 72 h (Fig. 5A). At the same time, the procyclic marker EP procyclin was clearly detectable by 24 h (Fig. 5A), and a little later the parasites resumed growth as procyclic forms (not shown). In the overexpression strain endogenous RBP10 persisted for the first 24 h, and no EP procyclin was seen. Even after tetracycline removal, when both RBP10 proteins disappeared, EP procyclin was not expressed (Fig. 5A) and indeed, the parasites died 1–3 days after the change of the medium (not shown). Again, addition of glucose to the procyclic medium didn't rescue the parasites (data not shown).
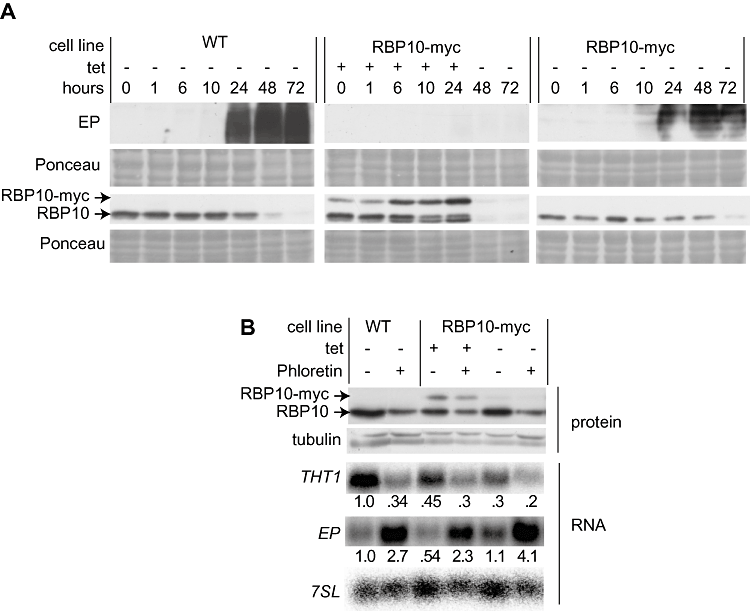
Effect of RBP10-myc expression on differentiation. A. RBP10-myc was inducibly expressed in differentiation-competent bloodstream-form trypanosomes (EATRO 1125), by addition of tetracycline to cells at a density of 2 × 105 ml−1. When cells attained a density of 1–2 × 106 ml−1, citrate and cis-aconitate were added and the culture was moved to a 27°C incubator. 24 h later, cells were shifted to procyclic medium. Western blots were probed for RBP10 and EP procyclin. Protein expression was assayed by Western blotting (approx 3 × 106 cells/lane). B. RBP10-myc can be overridden by phloretin. Cells as in (A) were grown with or without tetracycline. After 24 h, they were diluted to 2 × 105 ml−1 and allowed to grow for a further 24 h with continued tetracycline presence. Alternatively, they were diluted to 2 × 105 ml−1, and 100 µM phloretin was added (with continued tetracycline as required). After a further 24 h the cells were harvested and used for RNA (Northern blot) or protein detection. The experiment was done once with EATRO 1125 cells and once with Lister 427 cells, each with two technical replicates. The conclusion was always the same.
Targeted co-immunoprecipitation experiments revealed no evidence for a physical interaction between RBP10 and PTP1 and PIP39 (not shown) but as minor effects had been seen on the PIP39 mRNA, we investigated the effect of RBP10 alterations on expression of both phosphatases at the protein level. RBP10 RNAi in bloodstream forms, and RBP10-myc (over)expression, in either bloodstream or procyclic forms, had no effect on PTP1 or PIP39, as judged by Western blotting (not shown). Thus, there is no evidence that RBP10 exerts its effects on gene expression via PTP1-PIP39 signalling.
Finally, we asked whether RBP10 expression could prevent the response of bloodstream-form trypanosomes to phloretin, by adding phloretin 24 h after induction of expression of RBP10-myc. Phloretin still induced EP procyclin mRNA, and reduced THT1 mRNA (Fig. 5B). Thus glucose deprivation is able to induce these markers of differentiation even in the presence of RBP10. This suggests that the glucose response is either downstream of, or independent of, RBP10 action.
Detailed analysis of the RBP10 effects on THT1 expression
To investigate the effects of RBP10 alterations in detail, we focussed on the mRNAs encoding the glucose transporter THT1 and the phosphoglycerate kinase isozyme PGKC. Effects of RBP10 on transcription could not have caused the observed changes in either mRNA, because both are embedded in polycistronic transcription units, with neighbouring co-transcribed genes that are not co-regulated upon alterations in RBP10 expression. Nevertheless, we did check that RBP10 was indeed influencing THT1 mRNA degradation in bloodstream forms. We used Northern blotting to measure the levels of THT1 mRNA 15 min after treatment with 10 µg mol−1 Actinomycin D. In WT cells, about 50% of the mRNA remained, suggesting a half-life of around 15 min, whereas after RNAi induction, degradation was at least twice as fast (10% remaining after 15 min, data not shown). For PGKC, the half-life comparison was not possible after RNAi because the mRNA was too near the detection limit.
Expression THT1 and PGKC depends on specific sequences within the 3′ UTRs of the mRNAs (Blattner and Clayton, 1995; Hotz et al., 1995; Colasante et al., 2007). For this reason we cloned the intergenic regions of THT1 or PGKC downstream of a constitutively expressed CAT reporter (Fig. 6) and transfected the constructs into the RBP10 knock-down cell line. When RBP10 RNAi was induced, the amounts of CAT mRNA and CAT protein decreased (Fig. 6). This shows that RBP10 depletion affected 3′ UTR-mediated regulatory pathways.
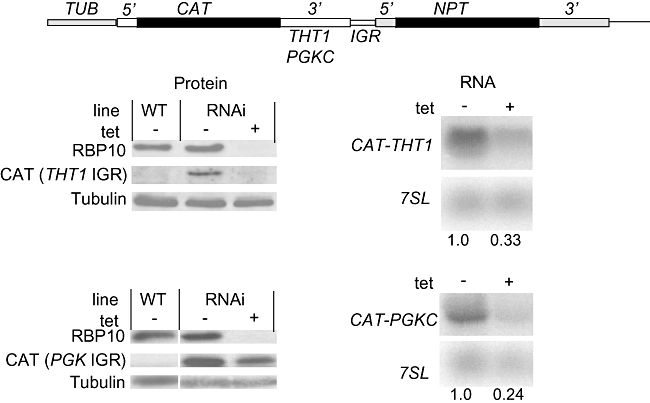
Effect of RBP10 depletion on expression of CAT reporter genes bearing wither a THT1 or PGKC 3′ UTR. The upper panel shows the plan of the plasmids; the lower panels are Western blots (left) and Northern blots (right) for one cell line for each reporter, with relative quantification underneath. By real-time PCR, the induction of RBP10 RNAi reduced the CAT-PGKC and CAT-THT1 mRNAs to about 40% and 30%, respectively, of the minus-tet control values.
We also asked whether the RRM alone was capable of inducing THT1 mRNA in procyclic forms. Various truncated versions of RBP10 were expressed and the level of THT1 mRNA was assessed (Fig. 7). As before, expression of full-length RBP10-myc resulted in a robust increase in THT1 mRNA (Fig. 7B, lane 3); the amount of RBP10 in the cells was at equivalent to that seen in bloodstream forms (Fig. 7C, compare lanes 1 and 3). The RRM alone was in contrast unable to upregulate THT1 (Fig. 7, lane 3), and deletion of either the N-terminal domain preceding the RRM (fragment F2, Fig. 7 lane 5) or part of the C-terminus (fragment F4, Fig. 7 lane 7) also abolished activity, despite good protein expression. Expression of the same protein fragments without the tag also had no effect (not shown). Thus several parts of RBP10 are required for its regulatory activity.
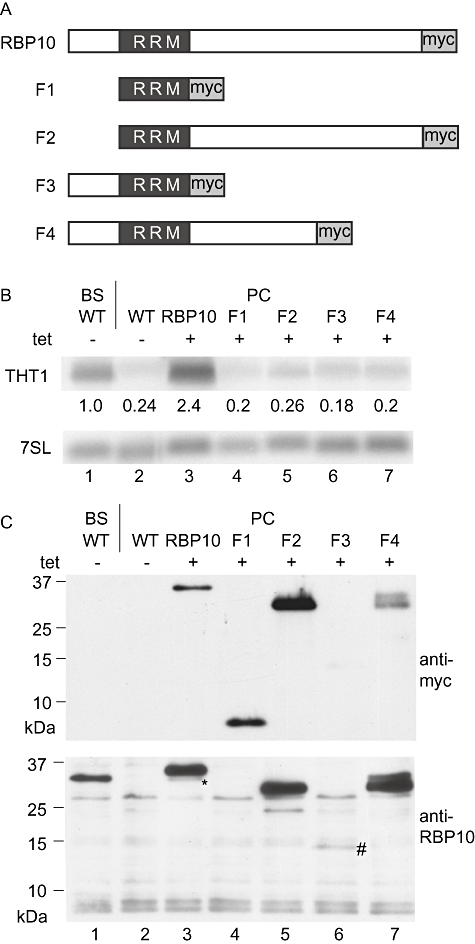
The N- and C-termini of RBP10 are required to increase expression of THT1 mRNA in procyclic forms. A. Fragments of RBP10 expressed. Sizes are: RBP10- 306 residues, 32 kDa; F1- 80 residues, 8 kDa; F2- 260 residues, 27 kDa; F3- 126 residues, 13 kDa; F4- 218 residues, 23 kDa. B. Levels of THT1 mRNA in procyclic trypanosomes expressing the different RBP10 fragments. C. Expression of the fragments in procyclic forms as determined by Western blotting using anti-myc (above) or anti-RBP10 (below) antiserum. A sample from bloodstream forms serves as a positive control (lane 1) and the background bands from the RBP10 antiserum serve as an internal loading control. F3 was very poorly expressed (lane 6, #), while F1 was not recognized by the anti-RBP10 antiserum (lane 4) and F4 appeared as two bands, each with the myc tag, suggesting some proteolytic degradation at the N-terminus. The asterisk indicates induced wild-type RBP10 in the RBP10-myc expressing cells (lane 3).
RBP10 has no detectable interaction with RNA
To investigate the identity of mRNAs bound to RPB10, we UV-irradiated cells expressing RBP10-myc, immunoprecipitated the protein, and subjected the bound RNA to high throughput cDNA sequencing. Very little RNA was obtained, and a comparison of the bound and unbound RNAs did not reveal any convincing RBP10-bound transcripts. Using reverse transcription and PCR, we looked for binding of potential targets such as THT1 and found no evidence that they were bound (not shown). Finally, we UV-irradiated cells in medium, immunoprecipitated RBP10-myc, digested unbound RNA, then end-labelled remaining RNA using 32P. Proteins with covalently bound RNA were then separated by SDS-PAGE and 32P was detected. This method detects RNA that is covalently bound to proteins during in vivo cross-linking (Hafner et al., 2010). A preparation made from cells expressing UBP1-myc (Hartmann et al., 2007) gave a clear signal at approximately the expected size (Fig. 8) but there was no signal whatsoever from the RBP10-myc preparation (Fig. 8). This was consistent with the absence of RBP10 in polysomal fractions (Fig. 1E). Similarly, although immunoprecipitation of RBP10-myc, or tandem affinity purification of TAP-RBP10, revealed a few possible protein interaction partners, none could be confirmed by reciprocal immunoprecipitation (Table S2 and not shown). Therefore, any interactions of RBP10 with proteins or RNA are likely to be either transient, or too weak to survive our purification procedures.
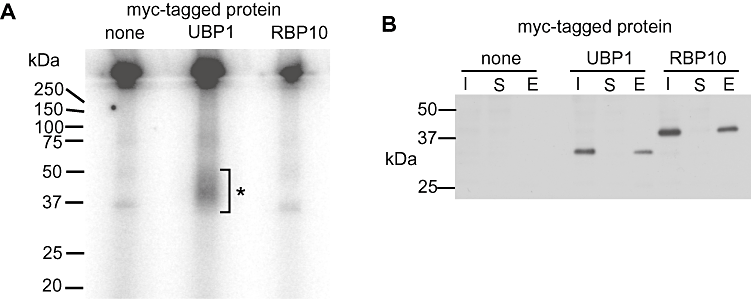
RBP10 does not appear to bind to RNA. Trypanosome cultures expressing myc-tagged RBP10 or UBP1, or with no myc tag, were irradiated at 254 nm. Proteins were precipitated with anti-myc antibody-coupled beads. RNA associated with the precipitating beads was partially digested, then remaining bead-associated RNA was 5′ labelled with 32P. Proteins were then separated by SDS-PAGE. About 60% of the immunoprecipitated and labelled sample was taken for phosphorimaging (A) and about 15%, at each stage, for Western blotting with anti-myc (B). In (B) I = input lysate; S = supernatant from the immunoprecipitation; E = eluate from the boiled beads; each sample represents approximately the same number of input trypanosomes. In (A) the labelled RNA-UBP1 is indicated by *; it always migrated as a smear, somewhat slower than the main protein band. The RBP10 lane was reproducibly identical to the control with no myc-tagged protein.
RBP10 can repress translation
Our experiments had already demonstrated that RBP10 was not stabilizing abundant mRNAs via direct binding. One alternative was that it destabilizes mRNAs. To assess this, we used a tethering assay. We first made a bloodstream-form cell line that constitutively expressed a CAT mRNA bearing the ‘B-box’ binding site for the bacteriophage lambda N protein (CAT-B-ACT) (Fig. 9A). The 3′ UTR was from the actin mRNA, which is not affected by RBP10 expression. As a control we used a cell line expressing CAT-ACT with no B-boxes (Fig. 9A). We then inducibly expressed fusion proteins bearing the lambda-N peptide at their N-termini: two controls, poly(A) binding protein (N-PABP1-myc) and N-GFP-myc, and N-RBP10 (Fig. 9A). As previously shown for procyclic forms (Delhi et al., 2011), expression of N-PABP1-myc in bloodstream trypanosomes carrying CAT-B-ACT caused a massive increase in CAT protein and RNA (Fig. 9D and E). A cell line expressing N-GFP-myc had less CAT than the starting CAT-B-ACT line. This is likely to be due to clonal variation, as a slight increase in CAT (not a further decrease) was seen after tetracycline induction (Fig. 9F). A positive effect of tethered GFP was also previously seen in procyclic forms (Delhi et al., 2011).
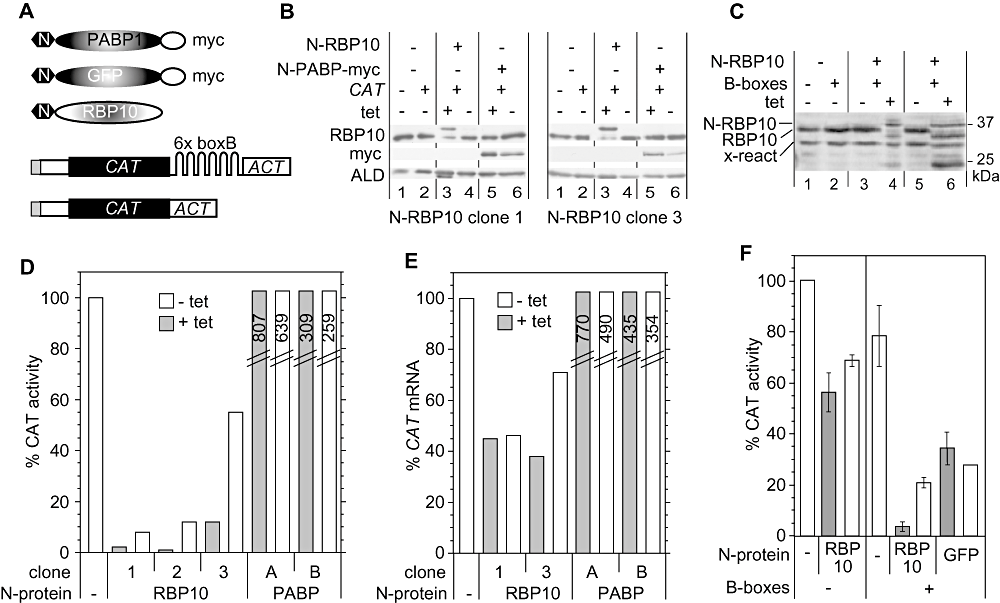
Tethering of RBP10 to an mRNA in bloodstream forms inhibits its translation. A. Schematic diagram of reporter mRNAs and tethered proteins, not to scale. The three expressed proteins (above) each bear an N-terminal lambda N peptide (hexagon), which is joined to PABP-myc, GFP-myc or RBP10. The two CAT reporters are shown below: the CAT gene is shown in black, spliced leader in grey and UTRs in white. B. Expression of lambda-N-RBP10 in clones 1 and 3. The Western blot was probed with anti-RBP10 and anti-myc, then re-probed for aldolase as a control. The aldolase protein runs very close to lambda-N-RBP10, which can still be seen on the left-hand ALD blot in track 3. C. Expression of lambda-N-RBP10 in a cell lines expressing CAT-ACT, compared with expression of lambda-N-RBP10 in clone 3. Some of the lambda-N-RBP10 has been degraded to a fragment of about 25 kDa. D. CAT activity in cell lines expressing lambda-N-RBP10 or lambda-N-PABP-myc. The activities were measured 24 h after tetracycline addition, and are expressed relative to a control line with no lambda-N protein. Results for PABP are for a single clone: ‘A’ is the experiment done with N-RBP10 clone 1, and ‘B’ the experiment done with N-RBP10 clones 2 and 3. The diagonal lines indicate that the PABP bars are not to scale; the values obtained are indicated within the bars. E. CAT mRNA levels in cell lines expressing lambda-N-RBP10 or lambda-N-PABP-myc. Levels were measured by Northern blotting 24 h after tetracycline addition, and were normalized to SRP or rRNA; they are expressed relative to a control line with no lambda-N protein. F. CAT activity in cells bearing either the CAT-ACT or the CAT-B-ACT reporter, with inducibly expressed lambda-N-RBP10 or lambda-N-GFP-myc. The assays were done twice for each cell line. The columns chow the arithmetic mean and the error bars show the range of values obtained. The CAT-B-ACT + lambda-N-GFP-myc cell line used was clone 3.
We now analysed three independent cloned cell lines co-expressing N-RBP10 and CAT-B-ACT, and one that co-expressed N-RBP10 and CAT-ACT. In all lines tested, weak expression of N-RBP10 was detected in the absence of tetracycline (Fig. 9B, lanes 4, Fig. 9C, lanes 3 and 5). Addition of tetracycline induced expression of N-RBP10 as expected, with similar levels seen in different clones (Fig. 9B, lanes 3, Fig. 9C, lanes 4 and 6). It was notable that in the tetracycline-treated lines, the level of untagged protein decreased (Fig. 9B, lanes 3) and degraded RBP10 appeared (Fig. 9C, lanes 4 and 6). We were worried that this decrease in wild-type RBP10 might by itself cause non-specific effects. Indeed, induced expression of N-RBP10 caused a slight decrease in cell growth (Fig. S1A), but upon pulsing with [35S]-methionine we saw no marked inhibition of translation (Fig. S1B). Probably, therefore, the tagged RBP10 is able to function similarly to native RBP10.
We now examined the effects of N-RBP10 expression in three cell lines with the CAT-B-ACT reporter. In the absence of tetracycline, each line had less CAT than the control line: this could have been due to clonal variation, or to leaky expression of N-RBP10, or a combination of the two. In each line, moreover, addition of tetracycline reduced CAT activity further – for lines 1 and 2, to almost undetectable levels, and fourfold for line 3 (Fig. 9D). The amounts of CAT mRNA were also decreased in the presence of N-RBP10, but the effect was not nearly as large (Fig. 9E). For example, in line 1 after tetracycline addition, CAT mRNA was 40% of the control lacking tethered RBP10, whereas CAT activity was less than 5%. This suggested that tethering of RBP10 to the reporter was inhibiting translation.
Figure 9F shows the control experiment: expressing N-RBP10 in cells bearing the CAT-ACT control plasmid. In the absence of tetracycline, this line had 30% less CAT activity than the parent line without N-RBP10 (perhaps due to clonal variation), but induction of N-RBP10 had almost no influence on CAT expression. The control, N-RBP10 co-expressed with CAT-B-ACT, again strongly decreased CAT (Fig. 9E). We concluded that strong inhibition of CAT expression depended upon binding of N-RBP10 to the reporter RNA.
Discussion
In this paper we have shown that the presence of RBP10 is necessary for expression of many bloodstream-form-specific genes, and sufficient to induce their expression in procyclic forms. Depletion of RBP10 in bloodstream forms decreased a large number of bloodstream-form-specific transcripts, while expression of RBP10 in procyclic forms increased them. The level of RBP10 also affected expression of two reporters with bloodstream-form-specific 3′ UTRs, showing that RBP10 affects mRNA at the post-transcriptional level. Our initial hypothesis was that RBP10 binds to, and stabilizes, bloodstream-form-specific mRNAs: but this was contradicted by the evidence: not only was RBP10 undetectable in the heavy polysome fraction, but we were completely unable to detect any specifically bound RNA. In contrast, tethering of RBP10 to a reporter mRNA strongly suppressed translation, and halved the target mRNA abundance.
We do not know how RBP10 inhibits translation, and also do not know how it might be recruited to target RNAs. Translation inhibition implies interactions with translation factors: for example, RBP10 might prevent translation initiation by interacting with intiation factors and impairing other protein–protein interactions. This would be consistent with the complete absence of RBP10 on polysomes. Protein–protein interactions have been reported for several RRM-domain proteins (Maris et al., 2005). Also, in bloodstream forms, expression of tethered RBP10 caused a decrease in the amount of native RBP10, which suggests that RBP10 might depend for its stability upon interaction with some other molecule that is present in limiting amounts. The existence of a stable complex was, however, contradicted by the fact that purification of tagged RBP10 revealed no convincing interaction partners – although tagged RBP10 was clearly functional in procyclic forms. Similarly, to inhibit translation, RBP10 would need to associate with target RNAs, yet no bound RNA was detected. This would be easy to understand if binding of RBP10 were to result in rapid target mRNA degradation, but the results of the tethering experiments did not support this conjecture. The alternative is that RBP10 is recruited via other proteins that bind to RNAs. To understand this further, it will be necessary to use methods that detect transient interactions.
Before discussing the possible role of RBP10 in regulating trypanosome gene expression, it is important to consider whether the effects of RBP10 depletion in bloodstream forms, or forced expression in procyclics, could simply be a consequence of growth inhibition. To answer this, we can compare our microarray results with those seen after other growth-inhibitory treatments. In bloodstream forms, RNAi targeting DHH1 (Kramer et al., 2010) also abrogates developmental regulation, but DHH1 is known to regulate mRNA degradation and translation. In contrast, lethal depletion of either the RRM proteins UBP1 and UBP2, or another essential RNA binding protein, ZC3H11 (D. Droll, R. Queiroz and C. Clayton ZMBH, unpublished) did not affect developmentally regulated mRNAs. Using a limited microarray, Koumandou et al. (Koumandou et al., 2008) also detected few transcriptome changes in bloodstream forms subjected to a variety of insults. In procyclic forms, for RNAi targeting ZC3H20, the overlap in transcriptome effects was restricted to just three mRNAs: THT1, an HSP70 (Tb09.160.3090), and Tb927.8.7340 (Ling et al., 2011); and depletion of DRBD3 also affected a completely different list of open reading frames (Estévez, 2008). Thus, impending growth inhibition by itself is not sufficient to induce differentiation-specific transcriptome changes in either life cycle stage. Instead, additional criteria must be fulfilled. These clearly include the ability to continue enough transcription and RNA processing to allow for transcriptome remodelling, but presumably there are more specific requirements as well.
We speculate that in bloodstream forms, RBP10 represses translation of a limited number of mRNAs that are required for procyclic-form gene expression. Upon release of RBP10 repression, the protein products are made and unleash a cascade of events that ultimately leads to partial procyclic differentiation. If this is true, though, what might the targets be? Because RBP10 tethering caused a 2- to 3-fold decrease in the attached mRNA, targets might be found among those mRNAs that increase in abundance after RBP10 RNAi in bloodstream forms. Direct RBP10 targets might also be expected to have more abundant mRNAs in procyclic forms than in bloodstream forms, and to show a decrease in abundance upon induced expression of RBP10 in procyclics. The gene products could also be essential in procyclic and differentiating cells, but not in growing bloodstream forms. Unfortunately, however, mRNAs that are indirectly affected might well have similar characteristics, and if RBP10 acts mainly on translation, or its effects depend on other proteins as well, any one or all of these criteria may not be valid. Indeed, examination of the mRNAs that increase at least 1.5-fold after RBP10 RNAi revealed none that satisfied all of the other criteria, although several fulfilled one or two (Table S1). Considering only essentiality during differentiation and in procyclic forms, some interesting candidates include Tb927.8.6650 (DRBD12, two-RRM-domains), Tb09.244.266 (TPR repeat), Tb927.7.2670 (CCCH zinc finger protein ZC3H21) and Tb927.7.6340 (palmitoylated). Others include Tb927.10.8050 (a putative phosphatase); Tb927.8.7820 (SSF50249 nucleic acid binding, cold-shock domain); and ZC3H20. However, multiple other potential RNA binding protein mRNAs were also altered, so the details of the regulatory cascade are likely to be rather complicated. Moreover, RBP10 RNAi affected mRNAs encoding cis-aconitate transporters, so could influence signalling by that route: it increased PAD2, and decreased PAD1.
Glucose transport carries significant control over the glycolytic flux (Bakker et al., 1999) and a reduction of glycolytic flux of 30–50% (for example, through phloretin treatment) blocks growth and causes differentiation-like transcriptome changes (Haanstra et al., 2011). RBP10 RNAi decreased the amount of THT1 mRNA; assuming that THT1 protein was also affected the consequent reduction in glucose might be sufficient to explain many of the transcriptome changes, as well as the lethality of RBP10 RNAi. This would be compounded by dysregulation of PGKB and PGKC, which is also lethal (Blattner et al., 1998). RBP10 RNAi, however, had a much more widespread effect on the transcriptome than treatment with phloretin. This was not due to more severe growth inhibition by the RBP10 RNAi. Both treatments were for 24 h; RBP10 RNAi starts to inhibit growth after 24 h have elapsed, and at the concentration used, phloretin inhibits growth without killing the cells. The mere presence of RBP10 was not sufficient to prevent the partial differentiation that is induced by glucose starvation: after phloretin treatment, RBP10 mRNA was unaffected and the amount of RBP10 protein was decreased only marginally. Also, the inhibition of differentiation through forced expression of RBP10 could be overridden by addition of phloretin. These observations indicate that the glucose-deprivation switch lies downstream of RBP10 action.
In conclusion, the simplest interpretation of our data is that RBP10 inhibits the translation of mRNAs encoding regulators, which promote procyclic-type expression. We hypothesize that these regulators may in turn affect the translation or stability of other, developmentally regulated, mRNAs. In procyclic forms, the appearance of RBP10 could decrease the synthesis of regulators that destablize bloodstream-form mRNAs and thus allow the latter to accumulate; at the same time, decreases in regulators that stabilize procyclic-form mRNAs would cause the latter to decrease. Some of the effects could be direct, and some indirect. In bloodstream forms, indirect effects seem quite likely. For example, depletion of RBP10 could allow synthesis of procyclic-type regulators. If one of these were required for the stability of the THT1 mRNA, disappearance of the regulator would result in the observed decrease in THT1 expression and hence in glucose transport. This would cause glucose deprivation, which is known to decrease mRNAs needed for bloodstream-form-type energy metabolism; the cells then die from metabolic imbalances and inadequate ATP production.
Experimental procedures
Trypanosome culture and plasmids
For all experiments except those involving differentiation, the trypanosomes used in this study were derived from the Lister 427 strain expressing the tet repressor. For stable transfections, 2 × 107 cells were transfected by electroporation with 10–12 µg of Not 1 linearized plasmid. After addition of the drug the cells were diluted to obtain single clones. The induction of RNAi or overexpression was done 24 h before the cells were collected using 100 ng ml−1 tetracycline, in the absence of selecting antibiotics. During proliferation assays cells were diluted every day to 2 × 105 cells ml−1.
For differentiation, pleomorphic cells of the EATRO 1125 strain, expressing the tet repressor (Benz et al., 2011), were used. Where relevant, expression of tagged RBP10 was induced for 24 h before the start of differentiation induction. At time 0, when cells were at a density of 1.5 × 106 cells ml−1 or higher, 6 mM cis-aconitate was added and cells were shifted to 27°C (Queiroz et al., 2009). After 24 h, the cells were centrifuged at 2000 g for 10 min and suspended in preheated MEM medium without tetracycline or cis-aconitate.
All plasmids are listed in Table S3, and oligonucleotides used in Table S4.
Cell fractionation, Western blotting and immunofluorescence
Cell fractionation was performed as described in Haile et al. (2007) and polysomes were prepared as described in Djikeng et al. (2003). For standard Western blot analysis, 3 × 106 cells were harvested by centrifugation at 2000 g for 10 min, washed with 1 ml PBS and suspended in 13 µl 2× protein buffer. Blots were incubated with RBP10 antibody (1:500, α rat), EP repeat (Cedar Lane, 1:2000, α mouse), XRND (1:1000, α rabbit) and tubulin (1:2000, α mouse). Immunofluorescence was done as described in Haile et al. (2007), except that additionally, z-stacks were taken for 3D-deconvolution using a Wiener filter.
RNA preparation and Northern blotting
RNA was obtained from growing cells at maximal densities of 1.5 × 106 ml−1 (bloodstream form) or 3 × 106 ml−1 (procyclic form). Cells were centrifuged and immediately resuspended in PeqGold Trifast (Peqlab) or the first solution from the RNeasy mini kit (Qiagen). For Northern blot analysis 10 µg of total RNA was separated by formaldehyde gel electrophoresis, blotted onto a Nytran membrane (GE Healthcare) and hybridized with radioactive DNA probes (Prime-IT RmT Random Primer Labelling Kit, Stratagene). The signals were measured with a phosphorimager and normalized to the signal of the 7SL probe (signal recognition particle RNA).
Microarray analysis
cDNA synthesis and slide hybridizations were performed as described in Queiroz et al. (2009) with the following changes: 12 µg of total RNA were used per hybridization; and cDNA synthesis was done with 400 U RevertAid H Minus Reverse Transcriptase (Fermentas), the appropriate reaction buffer and 40 U Ribolock (Fermentas). The reaction was incubated for 2 h at 43°C. DNase treatment and cDNA purification were done as in Queiroz et al. (2009).
We analysed the data using ExpressConverter and MIDAS software (freely available at http://www.tm4.org). Files obtained from the scan were background-subtracted and transformed into.mev – files using ExpressConverter. Using MIDAS the signal intensities were normalized by locally weighted linear regression and duplicate spots on each slide were merged. Log2 transformed data were exported to SAM as described (Tusher et al., 2001). All RNAs with a change of 1.5-fold or higher and a P-value of ≤ 0.5 was considered to be significantly regulated. Results are available at GEOarchive under series number GSE29176, with individual access numbers GSM722098-108.
Tandem affinity purification and myc immunoprecipitation
A total of 5 × 109 cells expressing TAP-RBP10 were used for tandem affinity purification as previously described (Estévez et al., 2003), the proteins were separated by gel electrophoresis and then all parts of the gel were examined by mass spectrometry. Similarly, myc-tagged RBP10 was immunoprecipitated as described above, from 3 × 108 cells. Specificity was assessed by comparisons with an anti-myc immunoprecipitation from cells that did not express a myc tag, and also with the mass spectrometry results from many other TAP or control purifications.
To search for RNA targets, 4 × 108 cells expressing RBP10-myc were grown to a density of 1.2 × 106 cells ml−1, washed once in ice-cold PBS, UV cross-linked, centrifuged again and snap-frozen. All following steps were carried out at 4°C or on ice. Cells were lysed in 400 µl lysis buffer containing 10 mM NaCl, 10 mM TRIS pH 7.5, 0.1% Igepal, 8 mM Ribonucleoside-Vanadyl Complexes and 1000 U RNasin (Promega). Cell debris was pelleted at 17.000 g and supernatant was adjusted to 150 mM NaCl. Fifty microlitres of myc-agarose beads (Sigma) was added, and incubated for 1 h. Afterwards beads were separated by centrifugation and washed three times with cold PBS. Proteinase K (Sigma) was added and degradation of proteins occurred at room temperature for 15 min. Then RNA was extracted using the Trifast FL (Peqlab) according to manufacturer's protocol.
RNA binding assay
A total of 2 × 108 bloodstream-form cells inducibly expressing either myc-tagged TbUBP1, or myc-tagged TbRBP10, or no myc-tagged protein were harvested 24 h after induction by tetracycline (100 ng ml−1 of culture). All the cultures were taken at cell densities of about 1.0 × 106 ml−1, and irradiated uncovered in Petriplates (14.5 cm × 2 cm, 25 ml per plate) with 0.3 J cm−2 of 254 nm UV light in a Stratalinker 2400 (Stratagene) at room temperature. The cultures were swirled shortly for mixing and cross-linked again. Subsequently, the cells were pelleted at 4°C and washed once with 1 ml ice cold 10 mM PBS-glucose. The pellet was snap frozen in liquid nitrogen and either stored at −80°C or processed immediately for immunoprecipitation.
Immunoprecipitation was done according to the PAR-CLIP protocol (Hafner et al., 2010) with minor modifications: the pellets of UV cross-linked cells were resuspended in 250 µl NP40 lysis buffer instead of three cell pellet volumes; for immunoprecipitation anti-c-myc antibody coupled agarose beads were used (Bethyl laboratories, catalog # S190-104); the second RNase treatment was done with 10 U µl−1 of RNase T1 instead of 100 U µl−1. Dephosphorylation and labelling of the RNA molecules bound to the tagged proteins were done exactly as in the PAR-CLIP description. To elute the RNA–protein complexes, 50 µl agarose beads was resuspended in 50 µl SDS-PAGE loading buffer, and complexes were separated by 15% SDS-PAGE. Samples taken for Western blotting were: Input: 10 µl of 250 µl lysate; unbound fraction: 10 µl; bound fraction: 10 µl of beads suspended in polynucleotide kinase buffer, just before radioactive labelling.
RNA-protein tethering assay
The tethering assay was done as described in Delhi et al. (2011). The RBP10 open reading frame was cloned in frame, downstream of the lambda-N peptide coding sequence. This and other constructs (Delhi et al., 2011) were co-expressed with a reporter mRNA bearing lambda-N binding sites (‘B-boxes’) between the CAT open reading frame and the actin 3′ UTR (Fig. 8C and see Table S3), or with a control lacking the B-boxes. CAT assays were done as previously described (Clayton, 1999).
Acknowledgements
We thank Keith Matthews for antisera to PTP1 and PIP39, and Keith Gull for anti-tubulin. M.W. and B.J. were supported by the Deutsche Forschungsgemeinschaft, SFB544 and Cl112/14, R.Q. by a fellowship from the Deutsches Akademisches Austauschdienst, and C.K. by a fellowship from the HBIGS graduate school. We are grateful to the Pathogen Functional Genomics Resource Center for provision of the microarrays, and to Esteban Erben (ZMBH) for advising M.W. on the tethering. All experimental work and much of the writing was done by M.W., with the following exceptions: R.Q. did the first two microarray hybridizations, B.S. did the C.A.T. reporter experiments in Fig. 6, C.K. made the polysome gradients and B.J. did the PAR-CLIP experiments (Fig. 8). B.J. also did the tethering control, together with Claudia Helbig (ZMBH), and Aditi Singh (ZMBH) produced Fig. S1. C.C. supervised the work and wrote the paper with M.W.




