The generation of nisin variants with enhanced activity against specific Gram-positive pathogens
Summary
Nisin is the prototype of the lantibiotic group of antimicrobial peptides. It exhibits broad spectrum inhibition of Gram-positive bacteria including important food pathogens and clinically relevant antibiotic-resistant bacteria. Significantly, the gene-encoded nature of nisin means that it can be subjected to gene-based bioengineering to generate novel derivatives. Here, we take advantage of this to generate the largest bank of randomly mutated nisin derivatives reported to date, with the ultimate aim of identifying variants with enhanced bioactivity. This approach led to the identification of a nisin-producing strain with enhanced bioactivity against the mastitic pathogen Streptococcus agalactiae resulting from an amino acid change in the hinge region of the peptide (K22T). Prompted by this discovery, site-directed and site-saturation mutagenesis of the hinge region residues was employed, resulting in the identification of additional derivatives, most notably N20P, M21V and K22S, with enhanced bioactivity and specific activity against Gram-positive pathogens including Listeria monocytogenes and/or Staphylococcus aureus. The identification of these derivatives represents a major step forward in the bioengineering of nisin, and lantibiotics in general, and confirms that peptide engineering can deliver derivatives with enhanced antimicrobial activity against specific problematic spoilage and pathogenic microbes or against Gram-positive bacteria in general.
Introduction
Lantibiotics are gene-encoded, ribosomally synthesized derived peptides that have attracted widespread scientific attention in recent years not only as promising safe and natural food additives but also as potential chemotherapeutic agents. The original, and most intensively studied, lantibiotic, nisin, has a long record of safe use, is US Food and Drug Administration (FDA) approved (Delves-Broughton, 1990) and is one of only a few bacteriocins to have been applied commercially (Twomey et al., 2002). It has antibacterial activity against many Gram-positive bacteria including food-borne pathogens such as staphylococci, bacilli, clostridia and mycobacteria. Nisin is also active against the bacteria responsible for bovine mastitis, an inflammation of the udder that is both persistent and costly to treat, and for this reason has been incorporated into a number of commercial products that are used as an alternative treatment to antibiotics (Sears et al., 1992; Wu et al., 2007). Studies have revealed that nisin, and a number of other lantibiotics, use the membrane-bound peptidoglycan precursor lipid II as a docking molecule, a step that facilitates two bactericidal activities, the inhibition of peptidoglycan biosynthesis and membrane pore formation (Brotz et al., 1998; Breukink et al., 1999; Wiedemann et al., 2001; Bonelli et al., 2006). This dual activity of nisin is possible due to the presence of two-structural domains, located at the N- and C-termini respectively. The N-terminal domain, containing three post-translationally incorporated (β-methyl)lanthionine rings (rings A, B and C), is linked to the C-terminal rings (rings D and E) by a flexible region, or hinge, consisting of three amino acids (Asn20–Met21–Lys22; Fig. 1). It has been established that the A, B and C rings form a ‘cage’ that facilitates binding of the pyrophosphate moiety of lipid II, thus interfering with cell wall synthesis (Hsu et al., 2004). This binding enhances the ability of the C-terminal segment, containing rings D and E, to form pores in the cell membrane, resulting in the rapid efflux of ions and cytoplasmic solutes (Wiedemann et al., 2001).
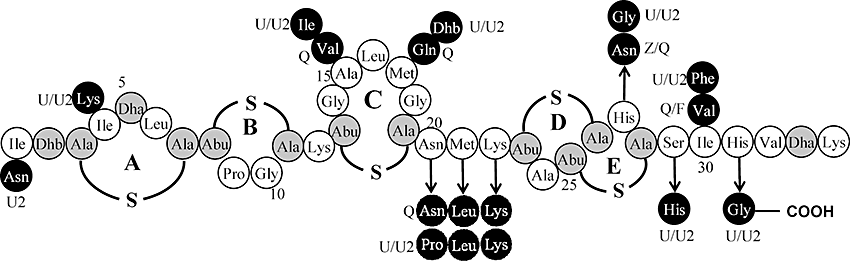
Structures of natural variants nisin A and nisin Z and putative structures of variant nisins Q, U and U2. Black circles indicate amino acid differences between the natural nisin variants. Post-translational modifications are indicated in grey. Dha, dehydroalanine; Dhb, dehydrobutyrine; Abu, 2-aminobutyric acid; Ala-S-Ala, lanthionine; Abu-S-Ala, 3-methyllanthionine.
To date, six natural forms of nisin have been identified. Nisin A (Kaletta and Entian, 1989), nisin Z (Mulders et al., 1991), nisin Q (Zendo et al., 2003) and nisin F (de Kwaadsteniet et al., 2008) are produced by Lactococcus lactis species while nisin U and nisin U2 are produced by Streptococcus uberis (Fig. 1) (Wirawan et al., 2006). The diversity of these natural variants highlights the extreme tolerance of certain residues and domains within the molecule to change. However, comparisons between closely (e.g. subtilin) and more distantly related (e.g. epidermin) lantibiotics also reveal that highly conserved elements, with essential structure/function roles, exist. Despite the relatively plastic nature of the nisin peptides, of the bioengineered derivatives of nisin that have been generated and characterized to date only two (T2S and M17Q/G18T) display increased activity against at least one Gram-positive bacterium, and even then activity is enhanced only with respect to a limited number of non-pathogenic indicator strains (Micrococcus flavus or Streptococcus thermophilus) (Kuipers et al., 1996; Cotter et al., 2005a; Lubelski et al., 2007). These, and indeed the majority of bioengineered peptides generated to date, have resulted from site-directed approaches, with random bioengineering of the intact nisin peptide having been carried out on a relatively small scale only (Spee et al., 1993). It is evident that random approaches can be worthwhile as recently an alternative approach involving randomization of a fragment, consisting of the N-terminal domain of nisin, yielded interesting results including enhanced IC50s against specific indicator strains (Rink et al., 2007a). To fully profit from the flexibility of nisin, we have generated the largest bank of randomly mutated nisin peptides reported to date and screened for enhanced antipathogen activity. This approach, especially when coupled to subsequent site-directed and site-specific saturation mutagenesis, yielded a significant amount of data with respect to the consequences of the alteration of specific nisin residues, and those within the hinge region in particular. The most significant outcome was the identification of several derivatives with enhanced activity against specific Gram-positive pathogens.
Results
Creation and screening of a bank of random nisin derivatives
A DNA fragment containing the nisA gene and its native Pnis promoter was amplified, and cloned into pPTPL (a reporter vector with a promoterless lacZ) to generate pDF03. This was subsequently introduced into L. lactis NZ9800. NZ9800 is derivative of a nisin-producing strain, L. lactis NZ9700, from which the nisA gene has been deleted (Table 1). The heterologous expression of nisA from pDF03 successfully restored nisin production to wild-type levels, confirming that this complementation system is suitable for expressing randomly mutagenized nisA genes. A second plasmid, pDF04 (pUC19-nisA), was used as a template for the generation of randomly mutated nisA fragments via mutazyme II PCR amplification (using conditions designed to achieve one nucleotide change on average per copy of nisA). These fragments were cloned into pPTPL and ultimately introduced into L. lactis NZ9800 as before. To increase the likelihood of identifying mutants with enhanced activity, a bank consisting of approximately 8000 potential variants was generated. In all cases the bioactivity of each variant was assessed by the size of the zone of inhibition in a deferred antagonism assay against L. lactis HP, Enterococcus faecium DPC1146 and Streptococcus agalactiae ATCC13813. Increased zone sizes could result from improved production, enhanced solubility or a higher specific activity, but these possibilities were not discriminated at this point, as strains with enhanced bioactivity are most likely to be industrially useful regardless of the underlying mechanism. It was established that approximately 20% of the blue colonies (indicative of effective Pnis promoter activity) tested displayed reduced bioactivity (smaller zones) as compared with the pDF03-containing control when assessed against the sensitive indicator strain L. lactis HP. Sequencing of a number of these revealed that mutations had occurred at the desired rate of between zero and three mutations per gene and at random locations throughout nisA including structural (e.g. I1F, T23I and S28F) and leader (e.g. R-1C and G-5D) regions. These initial results confirmed the creation of a bank of randomly altered nisA genes. At this stage, because the main focus of the study was the identification of mutants with enhanced activity, a decision was made to exclude colonies exhibiting a greatly reduced, or lack of, activity (in addition to those that had a white/light blue appearance on GM17-Xgal suggesting promoter mutations) from subsequent investigations. Of the remaining clones that were screened, all, bar one, of the mutant colonies tested exhibited either wild-type or reduced bioactivities against the three indicator strains (data not shown). The exceptional strain exhibited an enhanced bioactivity (approximately 50% increase in zone size compared with the corresponding positive control) against S. agalactiae ATCC13813 (Fig. 2A). Colony mass spectrometry (CMS) revealed that the peptide produced differed by 27 amu from wild-type nisin A. DNA sequence analysis revealed a C to A point mutation (AAA to ACA) resulting in a lysine22 to threonine (K22T) change within the hinge region of the mature nisin A molecule. This change is consistent with the Δ27 amu difference identified by CMS.
| Strain/plasmids | Relevant characteristics | Reference |
|---|---|---|
| Strains | ||
| L. lactis | ||
| L. lactis NZ9700 | Wild-type nisin producer | Kuipers et al. (1993; 1998) |
| L. lactis NZ9800 | L. lactis NZ9700 ΔnisA | Kuipers et al. (1993; 1998) |
| L. lactis NZ9800 pVE6007 | L. lactis NZ9800 harbouring pVE6007 | This study |
| L. lactis NZ9800 pPTPL | L. lactis NZ9800 harbouring pPTPL | This study |
| E. coli | ||
| E. coli MC1000 | E. coli host for pPTPL | O'Driscoll et al. (2004) |
| E. coli MC1000 pPTPL | E. coli harbouring pPTPL | O'Driscoll et al. (2004) |
| E. coli EC101 | E. coli host for pORI280 | Law et al. (1995) |
| E. coli Top10 | Intermediate cloning host | Stratagene |
| Indicator organisms | ||
| L. lactis ssp. cremoris HP | Nisin-sensitive indicator | UCC Culture Collection |
| E. faecium DPRC1146 | Nisin-sensitive indicator | DPRC Collection |
| S. agalactiae ATCC13813 | Nisin-sensitive indicator | American Type Culture Collection |
| S. aureus DPRC5245 | Nisin-sensitive indicator | DPRC Collection |
| S. aureus ST528(MRSA) | Nisin-sensitive indicator (MRSA) | BSAC |
| L. monocytogenes 10403S | Nisin-sensitive indicator | UCC Culture Collection |
| L. monocytogenes EGDe | Nisin-sensitive indicator | UCC Culture Collection |
| Plasmids | ||
| pCI372 | CmR; high-copy cloning vector | Hayes et al. (1990) |
| pDF05 | pCI372 with nisA under its own promoter | This study |
| pPTPL | TetR; lacZ; low-copy cloning vector | O'Driscoll et al. (2004) |
| pDF03 | pPTPL with nisA under its own promoter | This study |
| pUC19 | AMPR; lacZ; high-copy cloning vector. | |
| pDF04 | pUC19 with nisA under its own promoter | This study |
| pORI280 | RepA-, LacZ+ | Leenhouts et al. (1996) |
| pDF06 | pORI280 with nisA | This study |
| pDF07 | pORI280 with nisA-K22S | This study |
| pVe6007 | CmR; temperature-sensitive | Maguin et al. (1992) |
- DPRC, Dairy Products Research Centre, Moorepark; BSAC, British Society for Antimicrobial Chemotherapy.
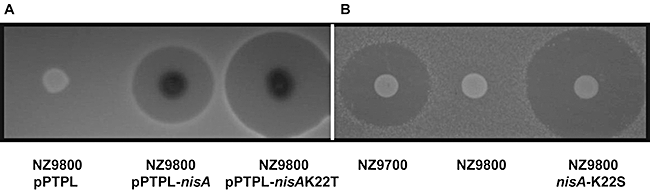
Growth inhibition of S. agalactiae ATCC13813 by the nisin A producing strains NZ9800pPTPL-nisA and NZ9700 and by the mutants K22T expressed in trans from the plasmid pPTPL and K22S expressed from a chromosomal replacement.
Creation of a K22S nisin mutant
It is notable that the K22T change introduces a hydroxylated residue which could act as a substrate for the lanthionine modification machinery (the five threonines in the native pro-peptide are all modified to dehydrobutyrines or β-methyllanthionines); CMS reveals that such modifications do not occur in this instance. To determine whether the beneficial consequences of the K22T change were specifically due to the introduction of a threonine or whether any hydroxylated amino acid would suffice, site-directed mutagenesis was undertaken to generate a K22S equivalent. The nisAK22S gene (generated by PCR-based mutagenesis) was inserted at the appropriate location in the L. lactis NZ9800 chromosome via double-cross-over recombination to generate L. lactis NZ9800::nisA-K22S. Results of deferred antagonism assays with S. agalactiae ATCC13813 established that bioactivity was not only restored but was in fact enhanced relative to that of L. lactis NZ9700 (Fig. 2B). CMS analysis confirmed the production of a peptide with a mass corresponding to a K22S change (3312 amu).
Creation and analysis of a bank of nisin hinge derivatives
As a consequence of the enhanced activities of K22T and K22S, coupled with previous observations highlighting the flexibility of the nisin hinge (Asn20, Met21 and Lys22) (Yuan et al., 2004), it was decided that a more in-depth investigation of this region should be carried out in the form of the most extensive mutagenesis of the nisin hinge to date. As our approach to random mutagenesis is not hinge specific, and because site-directed mutagenesis is relatively time-consuming, a saturation mutagenesis approach was undertaken to create a bank of nisin A hinge derivatives containing the vast majority of possible amino acid substitutions for each position. Although pPTPL was used successfully for random mutagenesis, its relatively large size of approximately 10.5 kb was considered unsuitable for the complete plasmid PCR amplification approach utilized for saturation mutagenesis. A smaller (approximately 6 kb) Escherichia coli–L. lactis shuttle vector pCI372 (Table 1) was considered to be potentially more useful. Its suitability with respect to the in trans expression of nisA was confirmed when, following the cloning of nisA into pCI372 to generate pDF05, its introduction into L. lactis NZ9800 was found to restore the nisin-positive phenotype. Saturation mutagenesis was performed on each codon using pDF05 and oligonucleotides (Table S1) that replace the specific codon with an NNK triplet, potentially encoding all 20 standard amino acids. Following complete plasmid amplification and introduction into the intermediate Top10 host, sequence analysis of a pooled bank of pDF05 derivatives confirmed randomization. Introduction of these variants into L. lactis NZ9800 allowed the expression of mutant nisin A peptides for further analysis. The bioactivity of approximately 150 L. lactis NZ9800 pDF05 derivatives was assessed for each of the three codons, again using deferred antagonism assays against several indicator organisms, including S. agalactiae ATCC13813, Staphylococcus aureus DPC5245 and S. aureus ST528 (a MRSA isolate) (Table 2). Further analyses in the form of CMS and gene sequencing were carried out to determine the extent of amino acid substitution at each position. The approach was highly successful in that of the 57 potential residue conversions that could have occurred (i.e. 19 at each position), 46 were isolated. In this instance all strains, regardless of bioactivity, were assessed with a view to comprehensively assessing the consequences of each individual hinge alteration. Given the multitude of derivatives generated, we will report the consequential impacts on relative bioactivity, as determined by deferred antagonism assays and calculated as zone size relative to the NZ9800 pCI372nisA, in the context of the nature (i.e. aromatic, charged, etc.) of the newly incorporated residue (Table 2).
| MRSA ST528 | S. aureus DPC5245 | S. agalactiae ATCC13813 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N20 | M21 | K22 | N20 | M21 | K22 | N20 | M21 | K22 | |
| N | 100 | 95 | X | 100 | 90 | X | 100 | 98 | X |
| Q | X | 92 | 82 | X | 94 | 77 | X | 98 | 82 |
| C | 0 | 0 | X | 0 | 0 | X | 0 | 0 | X |
| G | X | 125 | 118.5 | X | 123 | 112 | X | 115 | 126 |
| A | 60.5 | 132.5 | 126 | 69 | 135 | 117 | 52 | 105 | 137 |
| S | 83.5 | 105 | 154 | 87.5 | 110 | 124 | 72 | 110 | 142 |
| T | 79 | 110 | 146 | 90 | 112 | 145 | 46 | 110 | 153 |
| V | 68 | 135 | 107 | 77 | 156 | 113 | 63 | 101 | 126 |
| L | 43 | 76 | 93.5 | 69.5 | 70 | 89 | 69 | 58.5 | 108 |
| I | 28.5 | 107 | X | 49 | 115 | X | 61 | 92 | X |
| P | 125 | 40 | 90 | 123 | 32 | 92 | 22 | 0 | 65 |
| M | X | 100 | 77 | X | 100 | 79.5 | X | 100 | 104 |
| F | 11.25 | 20 | 29 | 20.5 | 21.5 | 56 | 15 | 0 | 57 |
| Y | 33.5 | 71 | X | 55 | 68 | X | 30.5 | 65 | X |
| W | 5.4 | 12 | 14.5 | 35.5 | 14 | 36 | 0 | 0 | 39 |
| D | 0 | X | 0 | 0 | X | 0 | 0 | X | 0 |
| E | X | 0 | 0 | X | 0 | 0 | X | 0 | 0 |
| R | 0 | 56.5 | 42.5 | 53.5 | 61 | 45 | 19 | 66 | 56 |
| H | 74.5 | X | 97 | 95 | X | 97 | 59.5 | X | 104 |
| K | X | 89 | 100 | X | 104 | 100 | X | 91 | 100 |
- Values are an average of duplicate experiments and represent zone size [diameter of zone minus diameter of bacterial growth (i.e. 5 mm) relative to that of the NZ9800 pCI372nisA control]. X = mutants not identified.
Incorporation of aromatic residues
An unusual feature of nisin is the absence of aromatic residues. To date all aromatic residue-containing forms of nisin have been bioengineered derivatives and all have displayed reduced antimicrobial activity [i.e. I1W, M17W, V32W, I30W, N20F and N20F/M21L/K22Q (Martin et al., 1996; Breukink et al., 1998; Yuan et al., 2004)]. Our results confirm this phenomenon in that the introduction of aromatic residues at any position in the hinge had a negative impact on nisin bioactivity, although this tended to vary depending on residue type and location. N20W, M21W and K22W all gave decreased zone sizes against S. agalactiae ATCC13813 (0–39%), S. aureus DPC5245 (14–36%) and S. aureus ST528 (5–14.5%) (Table 2). N20F, M21F and K22F changes also negatively impacted on the bioactivity of the producing strains against all indicators tested, although, on average, not to the same degree as tryptophan (ranging from 0% to 57%; Table 2). The bioactivity of NZ9800 N20Y resembled that of its N20F counterpart (N20Y slightly higher on average) while the M21Y equivalent was even less dramatically affected, i.e. it retained approximately 70% bioactivity against S. aureus strains and 65% bioactivity against S. agalactiae ATCC13813 (Table 2).
Consequences of the incorporation of charged residues
Nisin A is a cationic antimicrobial peptide due to the presence of five positively charged residues (Lys12, Lys22, Lys34, His27, His31) and the absence of negatively charged residues. The consequences of charge manipulation to date have been variable. In general the incorporation of negatively charged residues has had a very detrimental impact (e.g. the hinge mutants N20E, M21E and K22E; Yuan et al., 2004) whereas the introduction of positively charged residues has had less severe (V32K; Van Kraaij et al., 1997) and, in some cases, beneficial outcomes (with respect to anti-Gram-negative activity – N20K and M21K; Yuan et al., 2004). Our data confirm the detrimental impacts of introducing negatively charged residues into the hinge region in that N20D and K22D were devoid of bioactivity. A peptide corresponding to a K22D substitution could not be detected by CMS, indicating a negative impact on peptide production. It has previously been suggested that the introduction of a negatively charged residue at K22 in nisin Z may result in steric hindrance during the thioether bridge formation of ring D (Yuan et al., 2004). Similarly, N20D production is apparently reduced as a peptide of expected mass (3353.47 Da; Table S2) was only identified following small-scale purification prior to mass spectrometry analysis. Introduction of glutamic acid into the hinge region also resulted in loss of bioactivity for M21E, and a lack of any detectable production of a K22E peptide was consistent with previous findings (Yuan et al., 2004; Table 2).
Of the novel mutants in which a positive residue was introduced into the hinge, it was established that N20H and K22H displayed close to wild-type bioactivity levels (e.g. 95% and 97% retention of relative bioactivity, respectively, against S. aureus DPC5245; Table 2), whereas the introduction of arginine into the hinge (NZ9800 pCI372nisAN20R, M21R and K22R) resulted in greatly reduced bioactivities. It is thus apparent that although the introduction/exchange of positively charged residues within the hinge is generally tolerated, there are also structural considerations, with the bulkier arginine residues having the most negative influence.
Incorporation of hydrophobic residues
Various hydrophobic amino acids are naturally found within the hinge region of natural forms of nisin (i.e. nisin A, nisin Z – Met21; nisin Q – Leu21; nisin U/U2 – Pro20/Leu21; Fig. 1) suggesting that manipulations resulting in the interconversion or introduction of hydrophobic residues might be particularly successful. In general, the introduction of leucine (N20L, M21L, K22L), isoleucine (N20I, M21I) or methionine (K22M) resulted in the retention of relatively high levels of bioactivity, with the obvious exception of the N20I-producing strain which displayed particularly reduced activity against ST528 (Table 2). Notably, the bioactivity of the producer of M21L is somewhat decreased (58–76%). This observation is interesting as nisin Q, nisin U and nisin U2 naturally possess a leucine residue at this location, suggesting that this residue at this location may contribute to the variation in activity of natural variants. The consequences of the incorporation of valine residues varied very dramatically. As has been reported previously with respect to nisin Z (Yuan et al., 2004), it was apparent that a N20V strain exhibits reduced bioactivity levels (Table 2). In contrast the K22V-producing strain exhibited increased bioactivity levels against S. aureus DPC5245 (113%) and S. aureus ST528 (107%) and S. agalactiae ATCC13813 (126%; Table 2). The M21V strain was particularly notable in that, although wild-type bioactivity was apparent against L. lactis HP (data not shown) and S. agalactiae ATCC13813, an increase in relative bioactivity was evident with respect to S. aureus ST528 (135%) and DPC5245 (156%; Table 2 and Fig. 3). As a consequence of these observations, the M21V peptide was selected for purification and specific activity studies (see below). Proline substitutions also had very interesting consequences in that M21P and K22P strains had reduced relative bioactivity levels against all strains tested, an observation that is not unexpected given it has previously been established that proline incorporation, i.e. N20P/M21P, can result in the generation of a peptide incapable of pore formation (Wiedemann et al., 2001; Hasper et al., 2004). With respect to this double mutant, we can now report that it is the M21P change that is primarily responsible for the detrimental consequences because, in contrast, the single N20P strain displays enhanced bioactivity against S. aureus ST528 (125%), DPC5425 (123%) (Table 2, Fig. 3) and a number of other S. aureus strains tested (data not shown). In contrast, this strain displayed greatly reduced activity against the representative S. agalactiae strain, ATCC13813 (22%; Table 2). The strain variable nature of these results indicates that increases in bioactivity are not simply due to a general increase in production or enhanced rates of diffusion rate through the agar matrix, and must therefore be as a consequence of a greater specific activity against the target strain. For this reason N20P was one of three peptides selected for purification with a view to specific activity determination (see below).
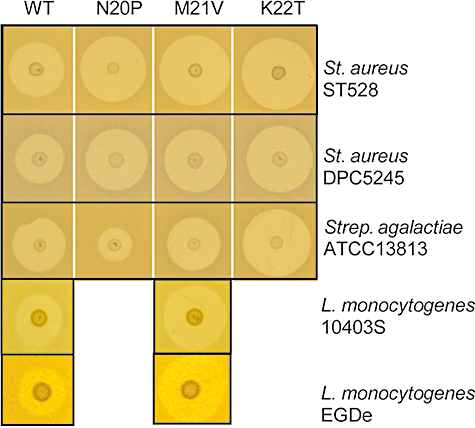
Growth inhibition of S. aureus ST528, S. aureus DPC5245 and S. agalactiae ATCC13813 by NZ9800pCI372nisAN20P, M21V and K22T and of L. monocytogenes 10403s and EGDe by NZ9800pCI372nisAM21V.
Incorporation of small and nucleophilic residues
A number of small amino acids have previously been introduced into the hinge region of nisin Z, including M21G (slightly reduced activity), K22G (slightly reduced activity) and N20A/K22G (as part of an epidermin-like hinge N20A/M21K/Dhb/K22G, greatly reduced activity; Yuan et al., 2004). We found that the consequence of making M21G and K22G changes to nisin A resulted in strains exhibiting a slightly increased relative bioactivity (in general, approximately 120% against S. aureus DPC5245 and ST528 and S. agalactiae ATCC13813; Table 2). This disparity could be as a consequence of indicator strain differences (in previous studies M. flavus and S. thermophilus were employed; Yuan et al., 2004), could represent a nisin A-specific phenomenon (previous studies having focused on nisin Z; Yuan et al., 2004) or could be as a consequence of relying on relative bioactivity rather than specific activity. Following site saturation mutagenesis, it was also now possible to assess the impact on bioactivity of the N20A alteration in isolation, which we found to result in decreased levels against all indicator strains (e.g. 60.5%, 52% and 69% against S. aureus ST528, S. agalactiae ATCC13813 and S. aureus DPC5245 respectively; Table 2). In contrast the other alanine-containing hinge mutants, M21A and K22A, had varying degrees of increased bioactivity (105–137%) against all the strains tested (S. aureus ST528, S. agalactiae ATCC13813 and S. aureus DPC5245; Table 2), therefore establishing that for positions 21 and 22 the presence of small amino acids can have a positive impact.
Incorporation of potentially modified residues
The most common post-translational modifications of lantibiotics involve the dehydration of serine to dehydroalanine (Dha) and of threonine to dehydrobutyrine (Dhb). These dehydrated residues interact with cysteine to form intramolecular lanthionine and β-methyllanthionine bridges respectively. Considering the key role of cysteine residues, it is perhaps unsurprising that the inclusion of additional cysteine residues generally impacts significantly on lantibiotic production and activity, i.e. the majority (8/11) of previously generated nisin Z derivatives incorporating cysteine are not produced (van Kraaij et al., 2000). Similarly, two strains generated in this study, N20C and M21C, did not produce a sufficient quantity of peptide to be detected by CMS analysis. However, small-scale purification revealed that small quantities of peptide were indeed produced and that the masses corresponded to N20C and M21C substitutions (Table S2). The activities of concentrated preparations of N20C and M21C were assessed and found to be drastically reduced against L. lactis HP and undetectable against S. agalactiae ATCC13813 and S. aureus DPC5245 relative to the wild-type peptide (data not shown).
The roles of serine and threonine residues in post-translational modification and their contribution to lantibiotic structure/function have been extensively investigated through site-directed approaches (Kuipers et al., 1992; Bierbaum et al., 1996; Wiedemann et al., 2001; Cotter et al., 2006). In general, any attempt to alter serines or threonines involved in (β-methyl)lanthionine formation impacts negatively on activity, although exchanging one for another is frequently tolerated (Kuipers et al., 1992; Rollema et al., 1995). Two mutants containing additional hydroxylated residues (K22T, K22S) have already been described in an earlier section. As both exhibit enhanced bioactivity against at least some target organisms, it was expected that mutants containing these alterations would also be isolated from the bank of site-saturated mutants. This proved to be the case as a number of mutants demonstrating enhanced activity against S. agalactiae ATCC13813 were found to contain K22T and K22S changes (Table 2, Fig. 3). It was apparent that the bioactivity of these strains was enhanced against S. aureus ST528 and S. aureus DPC5245 also (Table 2, Fig. 3). Of these the K22T peptide was selected as a representative for further investigations (see below). Strains producing N20T, N20S, M21T and M21S derivatives were also identified, all having either slightly increased, wild-type or decreased bioactivity (with the N20T producer being particularly poor against S. agalactiae ATCC13813; Table 2). Therefore while it is apparent that the incorporation of a hydroxylated residue into the hinge can have beneficial consequences, this is not universally the case. In all situations where a serine, threonine or cysteine residue was introduced, CMS indicated that the newly incorporated residue remained in an unmodified form (Table S2).
Incorporation of other residues
It has been reported that an N20Q substitution in nisin Z results in slightly diminished activity (Yuan et al., 2004). We can confirm that this is also true for the M21Q and K21Q substitutions (Table 2). With respect to asparagine, it was also established that an M21N mutant has approximately wild-type bioactivity against all strains tested (Table 2).
Anti-listerial activity of ‘hinge’ mutants
As a consequence of the risk associated with the survival and growth of Listeria monocytogenes in food, the bioactivity of the ‘hinge’ mutants against strains of this species was also subsequently assessed (data not shown). Of the mutants, it was consistently apparent that M21V displayed the greatest bioactivity against this pathogen (relative zone size –L. monocytogenes 10403S, 147%; L. monocytogenes EGDe, 153%) (Fig. 3), further justifying the selection of the corresponding peptide for specific investigation.
Specific activities of nisin A N20P, M21V and K22T
Given the rarity with which nisin derivatives with enhanced activity against Gram-positive bacteria have been identified, we investigated the basis for the enhanced relative bioactivity of three of the strains generated in this study (i.e. does increased zone size result from greater production and/or specific activity?). For this reason, with respect to each location within the hinge, peptide was purified from the strains exhibiting the greatest bioactivity, i.e. those producing N20P, M21V and K22T. Identical purification steps were employed to facilitate a comparison of production levels, which were found to be very similar in each case (Fig. 4A). Using equimolar concentrations of purified peptide, the specific activities of the wild-type nisin A, M21V, N20P and K22T peptides were determined (Fig. 4B). The data confirmed that the M21V peptide displays a 100% increased specific activity against S. aureus ST528, L. monocytogenes 10403S and L. monocytogenes EGDe but has wild type-like activity against S. agalactiae ATCC13813 (Fig. 3). The N20P peptide was also 100% more active than nisin A against S. aureus ST528 but was 75% less active against S. agalactiae ATCC13813. The enhanced activity (albeit strain specific) of N20P was surprising given the generally restrictive nature of proline residues with respect to conformational flexibility and its position within the ‘flexible’ hinge. However, the presence of a proline residue at position 20 of the natural variant nisin U is obviously tolerated and could contribute to its different target organism specificity (Wirawan et al., 2006). The K22T peptide possesses 100% greater specific activity against S. agalactiae ATCC13813 and S. aureus ST528 than its wild-type counterpart. This finding refutes the theory that the presence of a positively charged residue at position 22 (K22, H22) is required for the maintenance of a structure required for efficient pore formation (Yuan et al., 2004).
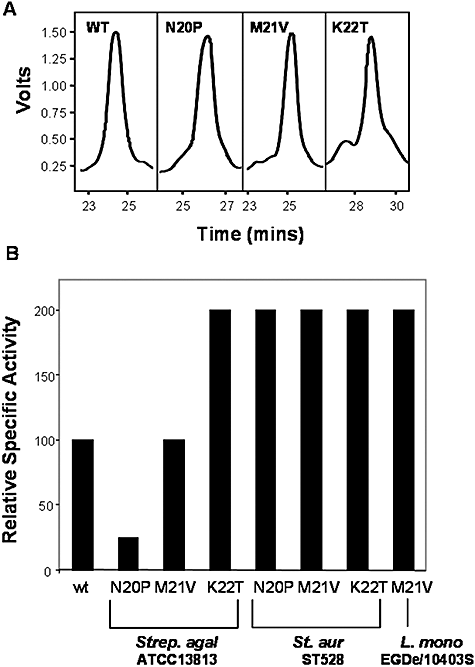
A. RP-HPLC of nisin A and derivatives thereof.B. Relative specific activity of purified nisin and nisin variants (with wild-type nisin at 100%).
Discussion
Following the initial site-directed mutagenesis of lantibiotics in the early 1990s (Kuipers et al., 1992; Liu and Hansen, 1992), the tolerance to change of regions of these peptides led researchers to speculate that these peptides may ultimately come to be regarded as primitive antibodies, potentially even containing essential and random domains (Liu and Hansen, 1992). The existence of such domains has become evident in recent years through comparison of the amino acid sequence of closely (and distantly) related lantibiotics (Cotter et al., 2005a), through structural analysis (Hsu et al., 2004) and through site-directed (Kuipers et al., 1996; Chatterjee et al., 2005; Lubelski et al., 2007) and alanine-scanning mutagenesis strategies (Cotter et al., 2006). The comparison between lantibiotics and antibodies is inaccurate in at least one sense, in that the producer does not normally generate a diverse population of these peptides (although it could be argued that a somewhat diverse population of lantibiotics exists as a consequence of evolution). It has been suggested that it may be possible to compensate for a strain's inability to randomize these peptides by devising ways to generate large populations of variants using genetic engineering or directed evolution approaches (Liu and Hansen, 1992). Despite this initial optimism, random and site saturation approaches have not been applied extensively to the bioengineering of lantibiotics or lantibiotic-derived peptides, with only a few notable exceptions (Kuipers et al., 1996; Field et al., 2007; Rink et al., 2007a). However, as positive outcomes have resulted from less extensive and more time-consuming site-directed mutagenesis strategies (for reviews see Cotter et al., 2005a; Lubelski et al., 2007), it was evident that the potential benefits may be realized in an extensive randomization approach.
Thus our initial goal was to create a bank of randomly mutated nisin derivatives, using an approach analogous to that previously successfully employed for the lantibiotic lacticin 3147 (Field et al., 2007). As the number of derivatives (approximately 8000) within the bank far exceeds that which has previously been generated, there was a greater likelihood that peptides with enhanced activity would be detected. As expected, a large number of strains were generated which exhibit a reduced, or lack of, activity. Although these were not investigated in great depth in this study, they will be the focus of future studies as establishing the tolerance of nisin to change at specific locations has the potential to reveal key residue(s) involved in structure/function relationships. These strains will also provide information that will guide subsequent bioengineering strategies with a view to generating more potent antimicrobial peptides. In this study, we set ourselves the more ambitious target of identifying nisin derivatives with enhanced bioactivity. While the identification of derivatives with enhanced anti-Gram-negative activity has been reported previously, the specific activity of these peptides was still below the range required to make clinical/commercial applications viable (Yuan et al., 2004). We decided to restrict our screen to find nisin derivatives that are enhanced with respect to the role for which nisin is most renowned, i.e. the inhibition of Gram-positive bacteria. There have been some rare successes resulting in enhanced activity against the non-pathogenic M. flavus and S. thermophilus. However, our focus was on the identification of derivatives with enhanced activity against strains of clinical or food relevance. Of the approximately 8000 mutants screened, one mutant (the K22T producer) was discovered that displayed enhanced antimicrobial activity against S. agalactiae ATCC13813, a pathogen associated with early perinatal human infections and bovine mastitis. The localization of this change within the hinge region of the peptide, coupled with previous site-directed mutagenesis data pertaining to this region, suggested this to be a somewhat flexible region (Yuan et al., 2004), and prompted further site-directed and site-saturation investigations that lead to the isolation of strains producing K22S, N20P and M21V and the associated peptides. While, upon initial investigation, these represent the strains/peptides of the greatest interest, the enhanced bioactivities of other strains, such as those producing M21A and K22A among others, suggest that these also merit closer inspection (including specific activity studies) in the future. This study has therefore tripled the total number of nisin mutants known to possess enhanced anti-Gram-positive activity.
While the identification of nisin derivatives with enhanced activity is in itself a rare event, the specific testing which establishes for the first time that nisin derivatives can possess enhanced activity against Gram-positive bacteria of clinical significance is especially noteworthy. The strain- and species-specific nature of this enhanced activity again raises the antibody analogy and, although it is unlikely that nisin derivatives will ever achieve a corresponding level of specificity, it would seem that the possibility exists that a selection of nisin derivatives might ultimately be generated, each dedicated to specific, distinct purposes. The antimicrobial activities of the four newly generated derivatives support this theory. More specifically, the enhanced activity of M21V nisin A against L. monocytogenes could make it a more preferable option than nisin A for some food biopreservation applications (Sobrino-Lopez and Martin-Belloso, 2008). This is particularly significant as L. monocytogenes are among the most naturally nisin-resistant Gram-positive pathogens. The similar production levels of M21V to wild-type nisin indicate that standard nisin purification/fermentation methods can be utilized, thus enabling its concentration and addition as a biopreservative analogous to nisin (e.g. Nisaplin®). The fact that such derivatives can be generated through the mere alteration of a single codon in situ in the producing strain, using the approach successfully employed to make a K22S derivative here, also means that the use of strains such producing bioengineered peptides may be accepted by food regulators as they do not involve the introduction of heterologous DNA and could be considered as ‘self-cloned’. Further studies will be crucial, however, to investigate the physical and chemical properties of M21V, including its solubility and stability at different temperatures and varying pHs before it can be used in food applications. From a veterinary perspective, K22T and N20P nisin A may have great potential in the treatment of bacteria that are responsible for bovine mastitis (staphylococci, but not streptococci, in the case of N20P). Nisin has been shown to be inhibitory to the principal Gram-positive mastitic pathogens (Broadbent et al., 1989) and as a result has been incorporated as the active ingredient in a number of commercial products that are used as an alternative treatment to antibiotics (Ross et al., 1999). The specific activities of N20P and K22T nisin A towards S. aureus and S. agalactiae, respectively, make them excellent candidates for the treatment of bovine mastitis. Similarly, the enhanced activity of N20P, M21V and K22T towards the MRSA strain ST528 is of particular note with implications for human biomedical applications. Antibiotic resistance among human pathogens has become a serious public threat; these enhanced nisin variants could become a welcome addition to the range of antimicrobials currently available for antibiotic-resistant Gram-positive infections. At the very least they could provide a valuable insight into structure/function relationships of the nisin molecule, in particular the species-specific nature of each nisin variant with its particular target organism, thereby facilitating the creation of further generations of nisin derivatives. Of course, each nisin derivative reported here could also form the basis of another round of ‘directed evolution’ to generate even more active peptides against suitable targets.
The success of the approach taken with respect to the creation and identification of nisin derivatives is in itself significant. In vivo based systems such as those employed here have several advantages including the availability of well-characterized expression hosts and cloning vectors that are capable of producing an almost infinite number of manipulated genes. However, a limitation associated with in vivo protein engineering is the abolition, or reduction in production, of novel structural variants as a result of the intolerance of the biosynthetic machinery to certain amino acid substitutions. Although novel in vivo and in vitro based systems have been developed to overcome this problem (Koponen et al., 2002; Xie et al., 2004; Kuipers et al., 2006; Rink et al., 2007b), it could be argued that the perceived limitations of minimal in vivo manipulations involving replacement of the structural gene alone are actually quite beneficial, i.e. when screening for novel variants, the most interesting will be those that are highly produced, do not overwhelm the associated lantibiotic immunity system, possess enhanced activity and can be produced in large quantities using standard purification/fermentation protocols. It would appear that a number of the derivatives described here satisfy these criteria.
Experimental procedures
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. L. lactis strains were grown in M17 broth (Oxoid) supplemented with 0.5% glucose (GM17) or GM17 agar at 30°C and GM17 supplemented with K2HPO4 (36 mM), KH2PO4 (13.2 mM), Sodium Citrate (1.7 mM), MgSO4 (0.4 mM), (NH4)2SO4 (6.8 mM) and 4.4% glycerol (GM17 freezing buffer) without aeration. E. coli was grown in Luria–Bertani broth with vigorous shaking or agar at 37°C. S. aureus strains were grown in Mueller–Hinton (MH) broth (Oxoid) or MH agar at 37°C, streptococci were grown in tryptic soy broth (TSB) or TSB agar at 37°C, Listeria strains were grown in brain–heart infusion (BHI) or BHI agar at 37°C. Antibiotics were used where indicated at the following concentrations: chloramphenicol and tetracycline at 5 and 10 μg ml−1, respectively, for L. lactis and at 20 and 10 μg ml−1, respectively, for E. coli. Erythromycin was used at 150 μg ml−1 and 5 μg ml−1 for E. coli and L. lactis respectively. Ampicillin was used at 100 μg ml−1for E. coli and X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was used at a concentration of 40 μg ml−1.
Random mutagenesis
DNA obtained from L. lactis NZ9700 (Hoffmann et al., 2004) was used as template for the amplification of a 372 bp fragment encompassing the nisA gene with KOD polymerase (Novagen) using the primers oDF101 and oDF102 (oligonucleotides utilized are listed in Table S1). PCR amplicons were purified using the QIAquick PCR purification kit (Qiagen), digested with BglII and XbaI (Roche) and cloned into similarly digested and Shrimp Alkaline Phosphatase (SAP; Roche) treated pPTPL. Following introduction into E. coli MC1000, plasmid was isolated from one clone and was sequenced (MWG Biotech, Germany) using the primer TETK P1 to ensure its integrity. The introduction of this plasmid, pDF03, into competent L. lactis NZ9800 successfully reinstated nisin activity. To provide sufficient quantities of template DNA for error-prone PCR (ep-PCR), nisA was re-amplified using pDF03 as template with KOD polymerase using the primers oDF101 and oDF103, digested with XbaI and EcoRI and cloned into similarly digested pUC19. Following introduction into E. coli Top 10 (Invitrogen), plasmid was isolated from one clone and was sequenced (MWG Biotech, Germany) using the primers M13FOR and M13REV to ensure its integrity. This plasmid, pDF04, was isolated from 100 ml of overnight culture using the Maxi-prep plasmid kit (Qiagen) to a concentration of approximately 1100 ng μl−1. pDF04 was used as template for the Genemorph II random mutagenesis kit (Stratagene) according to manufacturer's guidelines. To introduce an average of one base change in the 372 bp cloned fragment, amplification was performed in a 50 μl reaction containing approximately 500 ng of target DNA (pDF04), 2.5 units Mutazyme DNA polymerase, 1 mM dNTPs and 200 ng each of primers oDF101 and oDF102. The reaction was pre-heated at 96°C for 1 min, and then incubated for 22 cycles at 96°C for 1 min, 52°C for 1 min and 72°C for 1 min, and then finished by incubating at 72°C for 10 min. Amplified products were purified by gel extraction using the Qiaquick gel extraction kit (Qiagen), and re-amplified with KOD polymerase before being digested with BglII and XbaI (Roche), ligated with similarly digested and SAP-treated pPTPL and introduced into E. coli MC1000. To determine if the correct rate of mutation had been achieved recombinant plasmid DNA was isolated from selected clones using the QIAprep Spin miniprep kit (Qiagen) and sequenced (MWG Biotech). Transformants were pooled and stored in 80% glycerol at −20°C. Plasmid DNA isolated from the mutant bank was used to transform L. lactis NZ9800. Transformants (approximately 8000) were isolated from Q trays using the Genetix QPIX II-XT colony-picking robot and inoculated into 96-well plates containing GM17 freezing buffer, incubated overnight and subsequently stored at −20°C.
Site-directed mutagenesis
First, a 774 bp product encompassing approximately 300 bp either side of nisA was amplified with KOD polymerase using the primers oDF105 and oDF106, digested with EcoRI (Roche) and PstI (Roche) and ligated with similarly digested pORI280. Following transformation into E. coli EC101 (RepA+) plasmid was isolated from one clone (pDF06) and sequenced (MWG Biotech, Germany) using the primers pORI280FOR and pORI280REV to ensure its integrity. Mutagenesis of the nisA gene was achieved using a combination of the Quickchange site-directed mutagenesis strategy (Stratagene) and double-cross-over mutagenesis with pORI280 (RepA-, LacZ+) as described previously (Cotter et al., 2003; 2005b; 2006) using the Quickchange protocol as per manufacturers guidelines and using E. coli EC101 (RepA+) as host. To detect altered pORI280-nisA transformants, candidates were screened by PCR using a specific ‘check’ primer designed to amplify mutated plasmid template only and oDF106. Plasmid from one candidate (pDF07) was sequenced to verify the deliberate mutation and to confirm no other changes had been introduced. pDF07 was then introduced into NZ9800 pVe6007 by electroporation (Holo and Nes, 1995) and transformants were selected by growth on GM17-Ery-X-gal plates at 30°C. Integration of pDF07 by single-cross-over recombination and curing of the temperature-sensitive plasmid pVe6007 were achieved by growth at 37°C in GM17-Ery broth and plating on GM17-Ery-X-gal agar at the same temperature. Selected colonies were checked for their inability to grow on GM17-Cm agar at 30°C and then subcultured in GM17 at 37°C. Each subculture was spread on GM17-X-gal plates to identify candidates where pORI280 had excised and was lost (LacZ-) due to a second cross-over event. Mutant and wild-type revertants were distinguished by PCR using the check primer and oDF106 and also by deferred antagonism assay as candidate mutants exhibited a Bac+ phenotype and wild-type revertants a Bac- phenotype. Bac+ candidates were analysed by Mass Spectrometry to verify production of the mutant nisin peptide.
Saturation mutagenesis
To generate a template for mutagenesis, the 372 bp fragment encompassing the nisA gene was amplified with KOD polymerase using the primers oDF102 and oDF103, and was digested and subsequently cloned into pCI372. Following introduction into E. coli Top 10 cells, plasmid was isolated from one clone and was sequenced (MWG Biotech, Germany) using the primer pCI372REV to ensure its integrity. Saturation mutagenesis of the hinge region of nisA was carried out with pDF05 (pCI372-nisA) as template and using oligonucleotides containing an NNK codon in place of each native codon as listed in Table S1. PCR amplification was performed in a 50 μl reaction containing approximately 0.5 ng of target DNA (pDF05), 1 unit Phusion High-Fidelity DNA polymerase (Finnzymes, Finland), 1 mM dNTPs and 500 ng each of the appropriate forward and reverse oligonucleotide. The reaction was pre-heated at 98°C for 2 min, and then incubated for 29 cycles at 98°C for 30 s, 55°C for 15 s and 72°C for 3 min 30 s, and then finished by incubating at 72°C for 3 min 30 s. Amplified products were treated with Dpn1 (Stratagene) for 60 min at 37°C to digest template DNA and purified using the QIAquick PCR purification kit. Following transformation of E. coli Top 10 cells plasmid DNA was isolated and sequenced to verify that mutagenesis had taken place. Approximately 150 transformants encompassing each of the three hinge positions were chosen at random and inoculated into 96-well plates containing GM17 chloramphenicol, incubated overnight and stored at −20°C after addition of 80% glycerol.
Nisin purification
Lactococcus lactis NZ9700 or the mutant nisin strain of interest was subcultured twice in GM17 broth at 1% at 30°C before use. Two litres of modified TY broth was inoculated with the culture at 0.5% and incubated at 30°C overnight. The culture was centrifuged at 7000 r.p.m. for 15 min. The cell pellet was re-suspended in 300 ml of 70% isopropanol 0.1% trifluoroacetic acid (TFA) and stirred at room temperature for approximately 3 h. The cell debris was removed by centrifugation at 7000 r.p.m. for 15 min and the supernatant retained. The isopropanol was evaporated using a rotary evaporator (Buchi) and the sample pH adjusted to 4 before applying to a 10 g (60 ml) Varian C-18 Bond Elut Column (Varian, Harbor City, CA) pre-equilibrated with methanol and water. The columns were washed with 100 ml of 20% ethanol and the inhibitory activity was eluted in 100 ml of 70% IPA 0.1% TFA. Aliquots (15 ml) were concentrated to 2 ml through the removal of propan-2-ol by rotary evaporation. Aliquots (1.5 ml) were applied to a Phenomenex (Phenomenex, Cheshire, UK) C12 reverse phase (RP)-HPLC column (Jupiter 4u proteo 90 Å, 250 × 10.0 mm, 4 μm) previously equilibrated with 25% propan-2-ol, 0.1% TFA. The column was subsequently developed in a gradient of 30% propan-2-ol containing 0.1% TFA to 60% propan-2-ol containing 0.1% TFA from 10 to 45 min at a flow rate of 1.2 ml min−1.
Mass spectrometry
For CMS bacteria were collected with sterile plastic loops and mixed with 50 μl of 70% isopropanol adjusted to pH 2 with HCl. The suspension was vortexed, the cells were spun down in a benchtop centrifuge at 14 000 r.p.m. for 2 min and the supernatant was removed for analysis. Mass spectrometry in all cases was performed with an Axima CFR plus MALDI TOF mass spectrometer (Shimadzu Biotech, Manchester, UK). A 0.5 μl aliquot of matrix solution [alpha-cyano-4-hydroxy cinnamic acid (CHCA), 10 mg ml−1 in 50% acetonitrile-0.1% (v/v) TFA] was placed onto the target and left for 1–2 min before being removed. The residual solution was then air dried and the sample solution (re-suspended lyophilized powder or CMS supernatant) was positioned onto the pre-coated sample spot. Matrix solution (0.5 μl) was added to the sample and allowed to air dry. The sample was subsequently analysed in positive-ion reflectron mode.
Bioassays for antimicrobial activity
Deferred antagonism assays were performed by replicating strains on GM17 or GM17 X-gal agar plates and allowing them to grow overnight before overlaying with either GM17 or BHI or TS or MH agar (0.75% w/v agar) seeded with the appropriate indicator strain. Zone size was calculated as the diameter of the zone of clearing minus the diameter of bacterial growth (5 mm). For well-diffusion assays molten agar was cooled to 48°C and seeded with the appropriate indicator strain. The inoculated medium was rapidly transferred into sterile Petri plates in 50 ml volumes, allowed to solidify and dried. Wells (4.6 mm in diameter) were then made in the seeded plates. Purified nisin and mutant nisins were re-suspended in 0.005% acetic acid, serially diluted, 50 μl volumes were dispensed into the aforementioned wells and the plates incubated at 37°C overnight. Specific activity was calculated as the reciprocal of the highest dilution that gave a definite zone. Relative specific activity refers to the specific activity of each mutant expressed as a percentage of the wild-type nisin A value.
Acknowledgements
We are grateful to Evelyn Molloy for assistance in the screening of the random nisin mutant bank. This work was supported by the Irish Government under the National Development Plan, through a Science Foundation Ireland Investigator award to C.H., R.P.R. and P.D.C. (06/IN.1/B98), an Irish Research Council for Science Engineering and Technology (IRCSET) EMBARK postgraduate fellowship to D.F. and by an Enterprise Ireland Proof of Concept award.




