The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae
Summary
Maintenance of the intracellular homeostasis of metal ions is important for the virulence of many bacterial pathogens. Here, we demonstrate that the czcD gene of the human pathogen Streptococcus pneumoniae is involved in resistance against Zn2+, and that its transcription is induced by the transition-metal ions Zn2+, Co2+ and Ni2+. Upstream of czcD a gene was identified, encoding a novel TetR family regulator, SczA, that is responsible for the metal ion-dependent activation of czcD expression. Transcriptome analyses revealed that in a sczA mutant expression of czcD, a gene encoding a MerR-family transcriptional regulator and a gene encoding a zinc-containing alcohol dehydrogenase (adhB) were downregulated. Activation of the czcD promoter by SczA is shown to proceed by Zn2+-dependent binding of SczA to a conserved DNA motif. In the absence of Zn2+, SczA binds to a second site in the czcD promoter, thereby fully blocking czcD expression. This is the first example of a metalloregulatory protein belonging to the TetR family that has been described. The presence in S. pneumoniae of the Zn2+-resistance system characterized in this study might reflect the need for adjustment to a fluctuating Zn2+ pool encountered by this pathogen during infection of the human body.
Introduction
Metal ions are essential for the structure and function of many enzymes and regulatory proteins in bacteria. However, an excess of metal ions can be deleterious to the cell (Finney and O'Halloran, 2003). To maintain the homeostasis of metal ions, bacteria contain systems for metal ion uptake and efflux, of which the expression is tightly controlled by metal-responsive regulatory proteins (Hantke, 2001; Nies, 2003; Moore and Helmann, 2005; Pennella and Giedroc, 2005). Various systems involved in the uptake of metal ions have been studied in the human pathogen Streptococcus pneumoniae. Mn2+, and possibly also Zn2+, are taken up by the PsaBCA permease (Dintilhac et al., 1997; Lawrence et al., 1998; McAllister et al., 2004), whereas the adcCBA gene cluster likely encodes a Zn2+ ABC uptake system (Dintilhac et al., 1997; Dintilhac and Claverys, 1997). In addition, three iron uptake loci, piaABCD, piuBCDA and pit, have been described in S. pneumoniae (Brown et al., 2001; 2002). Both PsaBCA and AdcCBA, as well as their presumed substrates Mn2+ and Zn2+, are important for competence of S. pneumoniae (Dintilhac et al., 1997; Dintilhac and Claverys, 1997). Furthermore, PsaA (Berry and Paton, 1996; Johnston et al., 2004), PiaA and PiuA (Brown et al., 2001) have been shown to contribute to virulence, and moreover, PsaA contributes to oxidative stress resistance (Tseng et al., 2002; Johnston et al., 2004).
So far, three regulators have been described in S. pneumoniae, that regulate metal ion uptake and contribute to virulence. First, the DtxR-family regulator PsaR represses psaA and other genes in the presence of a high Mn2+ concentration (Johnston et al., 2006). Second, the orphan response regulator RitR (Throup et al., 2000) functions as a repressor of iron uptake via the piuABCD operon (Ulijasz et al., 2004). Third, TCS04 activates expression of psaBCA (McCluskey et al., 2004).
Next to metal ion uptake systems, bacterial genomes encode genes that enable the cell to cope with high concentrations of metal ions (Nies, 2003). Prominent protein families include: (i) the resistance nodulation family (RND), which are proton-driven antiporters found in all kingdoms of life; (ii) the cation diffusion facilitators (CDF family) driven by a chemiosmotic gradient or a potassium gradient (Haney et al., 2005); and (iii) P-type ATPases that are driven by ATP hydrolysis (Nies, 2003). In several bacteria, the CDF-family protein CzcD is known to be an important heavy-metal ion-resistance determinant. In the Gram-negative bacterium Ralstonia metallidurans CH34 (= Alcaligenes eutrophus), the czcRS two-component system is involved in regulation of the czcD and czcCBA genes, that mediate resistance against Co2+, Zn2+ and Cd2+ (Nies and Silver, 1989; van der Lelie et al., 1997). In Staphylococcus aureus CzrA, a member of the ArsR/SmtB family of DNA binding proteins, functions as a repressor of the czr operon, that consists of czrA and the gene encoding the CzcD homologue CzrB (Xiong and Jayaswal, 1998; Kuroda et al., 1999; Singh et al., 1999). CzrA-mediated repression is alleviated in the presence of Zn2+ and Co2+ (Xiong and Jayaswal, 1998; Kuroda et al., 1999; Singh et al., 1999). The Bacillus subtilis cation efflux pump czcD, which mediates resistance against Zn2+, Co2+, Ni2+ and Cu2+, is regulated by an ArsR-type repressor (CzrABS) as well (Moore et al., 2005).
As metal ions like Zn2+, Fe2+ and Cu2+ are also necessary for the proper functioning of the human immune system (Percival, 1998; Shankar and Prasad, 1998; Rink and Gabriel, 2000; Schaible and Kaufmann, 2004), they might have a significant influence on the interaction between S. pneumoniae and its host. Noteworthy, the concentration of Zn2+ may vary greatly between different sites in the human body (15 μM in serum versus 229 μM in the lungs) (Versieck, 1985). In the light of the pivotal role of CzcD homologues in metal ion homeostasis in other organisms, the function of czcD in S. pneumoniae was studied. In this pathogen we demonstrate that czcD is an important determinant for resistance against elevated levels of Zn2+. Furthermore, a novel TetR-family regulator, SczA, is shown to function as a Zn2+/Co2+/Ni2+-dependent transcriptional activator of czcD by binding to a regulatory cis-element in the czcD promoter, that is conserved in several related streptococci.
Results
Regulation and function of czcD in S. pneumoniae
The S. pneumoniae czcD orthologue is located in a possible operon with the genes spr1671, encoding a MerR-family regulator, and adhB, encoding a zinc-containing alcohol dehydrogenase, followed by a putative terminator (Fig. 1A). To be able to study the expression of the czcD operon in detail, an ectopic transcriptional lacZ fusion was constructed to the predicted promoter of czcD in D39 (Fig. 1A). Of various metal ions tested, namely Mn2+, Mg2+, Fe2+, Fe3+, Zn2+, Co2+, Ni2+ and Cu2+, only Zn2+, Co2+ and Ni2+ caused induction of PczcD–lacZ expression (Table 1). To investigate the physiological function of CzcD in S. pneumoniae, growth of an in-frame czcD deletion strain was compared with that of the wild-type in GM17 (Terzaghi and Sandine, 1975; a complex medium used for growth of streptococci, based on casein, soy peptone, beef extract and yeast extract) supplemented with various metal ions. This showed that the czcD mutant displays a strongly decreased resistance against Zn2+ compared with the wild-type (Fig. 2A). In addition, resistance against Co2+ was also lower in the czcD mutant (Fig. 2B), while resistance to Ni2+ was slightly higher compared with the wild-type (Fig. 2C). These data indicate that CzcD protects S. pneumoniae primarily against Zn2+ toxicity, and to a lesser extent against Co2+.
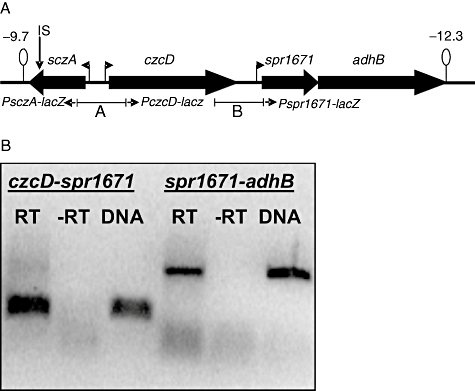
Organization of the czcD operon in S. pneumoniae.A. Schematic overview of the czcD locus in S. pneumoniae R6. Lollipops indicate terminator structures with the numbers indicating the predicted ΔG0 in kcal mol−1. Right- or left-pointing flags indicate promoters. Below the genes, fragments used for construction of the lacZ fusions to PsczA and PczcD (A) and Pspr1671 (B) in pPP2 are indicated. IS indicates the position of the mariner insertion in strain MP112.B. RT-PCR using primers spr1671-2/czcDKO-3 (czcD–spr1671 intergenic region) and spr1671-3/spr1671-4 (spr1671–adhB intergenic region). DNA, positive control PCR on D39 chromosomal DNA. ‘RT’ and ‘-RT’ indicate PCR on total RNA isolated from D39 wild-type grown in GM17 + 0.05 mM Co2+, with and without reverse transcription respectively.
| Medium | β-Galactosidase activity | ||
|---|---|---|---|
| wt | ΔsczA | ΔczcD | |
| GM17 | 3 (2) | 3 (2) | 12 (3) |
| Zn2+ (0.1 mM) | 66 (4) | 3 (2) | 85 (10) |
| Co2+ (0.05 mM) | 78 (5) | 5 (2) | 25 (8) |
| Ni2+ (0.2 mM) | 160 (16) | 6 (2) | 52 (9) |
- β-Galactosidase activity (Miller Units) is given for strains MP103 [D39 ΔbgaA::PczcD–lacZ, wt (wild-type)] and MP107 (D39 ΔsczAΔbgaA::PczcD–lacZ, ΔsczA) and MP111 (D39 ΔczcDΔbgaA::PczcD–lacZ, ΔczcD) grown in GM17 with the indicated metal ions. Standard deviation of three independent experiments is given between brackets.
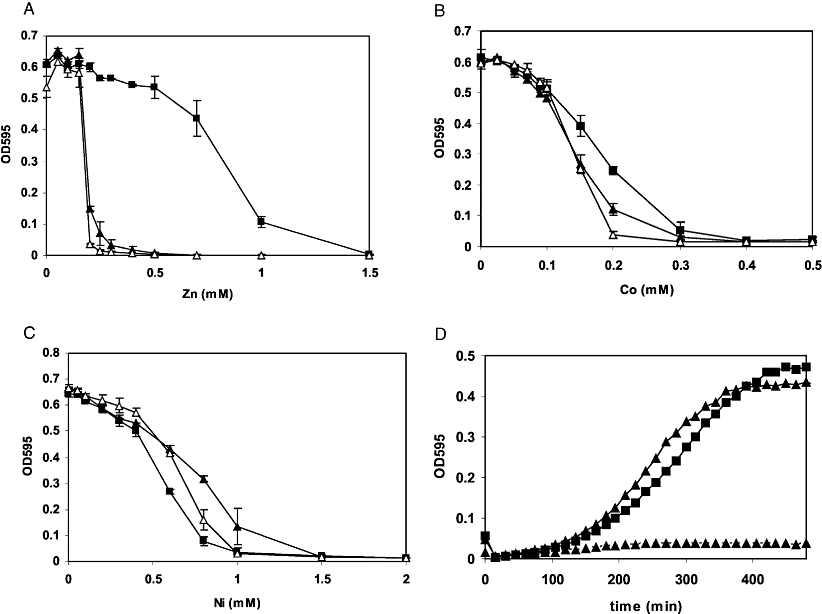
Optical densities at 595 nm (OD595) after 8 h of growth of D39 (black squares), MP102 (D39 ΔczcD, white triangles) and MP100 (D39 ΔsczA, black triangles) in GM17 with increasing concentrations of Zn2+ (A), Co2+ (B) or Ni2+ (C). Averages and standard deviations of three experiments are shown.D. Growth of MP101 (D39nisRKΔsczA) containing H6-sczA under control of the nisin promoter (pMP6, triangles) or the empty vector (pNZ8048, triangles, discontinuous line) and D39nisRK containing pNZ8048 (squares) in GM17 with 0.3 mM Zn2+ and 2 ng ml−1 nisin.
In R. metallidurans, deletion of czcD resulted in increased transcription of the czcCBA heavy-metal-resistance genes, likely due to an increase in the intracellular concentration of metal cations (Nies, 1992; Anton et al., 1999; Grosse et al., 2004). To investigate the possibility that in S. pneumoniae CzcD has a similar effect on expression of its own gene, we measured expression of the PczcD–lacZ transcriptional fusion in the czcD-deletion mutant (Table 1). In the absence of added metal ions, there was expression from PczcD in the mutant but not in the wild-type. The induction of expression by Zn2+ was slightly higher in the czcD mutant than in the wild-type. Surprisingly, however, expression from the czcD promoter (PczcD) in response to Co2+ and Ni2+ was threefold lower in the czcD mutant compared with the wild-type. Thus, deletion of czcD interferes with the responsiveness of PczcD to Co2+ and Ni2+.
Identification of SczA, a novel TetR-family regulator involved in activation of czcD
blast searches revealed that the S. pneumoniae genome does not contain orthologues of the ArsR-type repressor czrA that regulates expression of czcD in both B. subtilis and S. aureus (Xiong and Jayaswal, 1998; Singh et al., 1999; Kuroda et al., 1999; Moore et al., 2005). Therefore, we used strain MP103 (D39 ΔbgaA::PczcD–lacZ) to perform random mutagenesis with the Himar1 MarC9 transposon (Lampe et al., 1996), screening for mutants that are disturbed in the Zn2+-dependent induction of PczcD–lacZ expression. Random mutants were selected in the presence of a subinhibitory concentration of 0.25 mM Zn2+ and X-gal, a condition that normally leads to expression of PczcD–lacZ and thus blue colonies. Five white colonies were found among 12 500 blue colony-forming units. The transposon insertion sites were determined to be all at the same position in spr1673, encoding a TetR-family regulator (Ramos et al., 2005), which lies upstream of czcD (Fig. 1A). To unambiguously prove that spr1673 is involved in activation of czcD, we replaced the gene with a spectinomycin-resistance marker. In the resulting mutant, czcD expression was no longer induced by Co2+, Zn2+ or Ni2+ (Table 1). We propose the name SczA for the newly identified regulator, which stands for streptococcal czcDactivator. Thus, SczA activates transcription of the czcD gene in S. pneumoniae, in response to Co2+, Zn2+ or Ni2+.
As SczA activates expression of czcD, we hypothesized that the sczA mutant has the same phenotype as the czcD mutant. This is indeed the case (Fig. 2A–C). In trans expression of a his-tagged SczA version complemented the Zn2+-sensitive phenotype of the sczA mutant, excluding the possibility of polar effects of the sczA deletion on czcD (Fig. 2D).
Microarray analysis of the sczA mutant
To determine the influence of the sczA mutation on the transcriptome of S. pneumoniae D39, the wild-type was compared with the isogenic sczA mutant by use of DNA microarrays. To exclude effects due to the increased toxicity of Zn2+ to the sczA mutant compared with the wild-type, strains were grown in GM17 with 0.05 mM Co2+. Under these conditions, expression from PczcD is activated by SczA, and growth of the mutant and the wild-type is comparable (Fig. 2). The transcriptome analysis showed that in addition to czcD, also two downstream genes, namely the MerR-family regulator spr1671 and the zinc-containing alcohol dehydrogenase adhB (spr1670), were strongly downregulated in the mutant (Table 2).
| Gene namea | Functionb | Ratioc |
|---|---|---|
| SP0202/spr0183 | Anaerobic ribonucleoside-triphosphate reductase NrdD | 2.7 |
| SP0203 | Hypothetical protein | 2.5 |
| SP0204/spr0184 | Predicted acetyltransferase, GNAT family | 2.6 |
| SP0205/spr0185 | Anaerobic ribonucleoside-triphosphate reductase activating protein NrdG | 2.5 |
| SP0206 | Hypothetical protein; uridine kinase | 2.6 |
| SP0207/spr0186 | Hypothetical protein; uridine kinase | 2.6 |
| SP1855/spr1670 | Alcohol dehydrogenase, zinc-containing, AdhB | −3.9 |
| SP1856/spr1671 | Transcriptional regulator, MerR family | −3.8 |
| SP1857/spr1672 | Cation efflux system protein CzcD | −17.5 |
| SP1858/spr1673 | TetR-family transcriptional regulator protein SczA | −6.2 |
- a. Gene numbers refer to TIGR4 and R6 locus tags.
- b. TIGR annotation/R6 annotation (Hoskins et al., 2001; Tettelin et al., 2001).
- c. Ratios > 2.0 or < −2.0 (signal intensity for MP100 divided by that for D39), which have a Bayesian P-value < 0.001 and a false discovery rate < 0.01.
There is a 243 bp intergenic region between czcD and spr1671, but no intergenic region between spr1671 and adhB (Fig. 1A). Thus, spr1671 and adhB are likely to be regulated by SczA either via PczcD or via a putative promoter just upstream of spr1671. To investigate this, a transcriptional lacZ fusion with the czcD–spr1671 intergenic region was constructed (Fig. 1A). This lacZ fusion displayed 7 Miller Units promoter activity, which was neither influenced by the sczA mutation nor by the addition of metal ions to the medium (data not shown). In addition, reverse transcription polymerase chain reaction (RT-PCR) showed that spr1671 and adhB are located on the same transcript as czcD (Fig. 1B). Thus, we conclude that spr1671 and adhB are regulated by SczA via PczcD and are at least partially co-transcribed with czcD, i.e. form an operon with it.
A transcriptional unit (spr0183–spr0186) containing the class III nucleotide reductase encoding genes nrdD and nrdG, involved in ribonucleoside triphosphate synthesis, was upregulated in the sczA mutant, suggesting that SczA is directly or indirectly a repressor of these genes. Using a lacZ fusion to the promoter of nrdD (spr0183), the array data could be validated (Table 3). However, expression of PnrdD also seems to be affected by another factor, as an effect of SczA was only seen in the presence of Co2+. In conclusion, under these experimental conditions, SczA influences the expression of only a limited number of genes.
| Medium | β-Galactosidase activity | |
|---|---|---|
| wt | ΔsczA | |
| GM17 | 18 (3) | 21 (3) |
| Co2+ (0.05 mM) | 32 (2) | 103 (5) |
- Standard deviation of three independent experiments is given between brackets.
The sczA–czcD genomic organization is conserved among several streptococcal species
Using blast searches, several organisms were found to contain an orthologue of SczA. Only in S. mitis (90% identity), S. thermophilus (73% identity), S. pyogenes (50% identity) and S. agalactiae (48% identity), which contain the closest SczA homologues, the putative sczA genes were located immediately next to a czcD orthologue (data not shown). This suggests that in these streptococci, SzcA has a similar regulatory function of czcD expression as we here found in S. pneumoniae.
As the sczA–czcD gene order is conserved in the above-mentioned streptococci, the sczA–czcD intergenic regions from these five organisms were subjected to the online tool Gibbs Motif Sampler (Thijs et al., 2002). This resulted in the identification of a conserved palindromic sequence, which is present in the sczA–czcD intergenic region of each of the five organisms, and of several conserved residues that are present just upstream of this palindrome (Fig. 3). Searching the entire S. pneumoniae R6 and D39 genomes for the presence of the putative SczA operator using Genome2D (Baerends et al., 2004) did not reveal additional promoter regions containing a similar sequence.
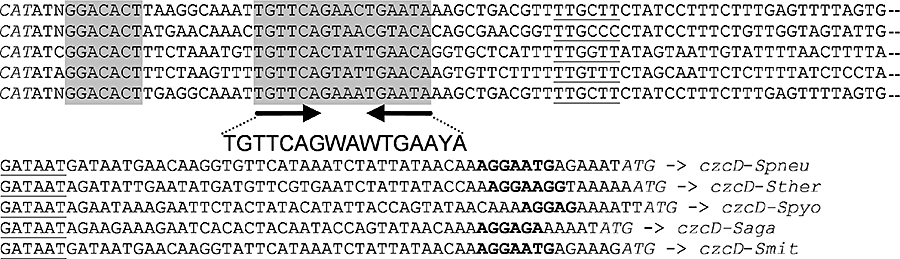
Identification of a SczA operator sequence in PczcD. Position of putative SczA operators (grey shading) in PczcD of S. pneumoniae R6 (Spneu), S. pyogenes MGAS5005 (Spyo), S. agalactiae A9 (Saga), S. thermophilus CNRZ1066 (Sther) and S. mitis NCTC 12261 (Smit). The palindrome is indicated with inverted arrows (→←). The consensus sequence is given below the alignment. W = A or T, Y = C or T. Putative core promoter sequences are underlined. Ribosome binding sites are in bold face. Start codons of czcD and sczA are in italic type. N, inserted base to align sequences.
Identification and verification of a SczA operators in PczcD
To find out whether the identified conserved DNA motif (hereafter designated as motif 1, 4, 8) mediates the SczA-dependent transcriptional control of PczcD, several lacZ fusions were constructed, in which PczcD was truncated from the 5′ end (Fig. 4). This showed that removal of the few conserved bases upstream of the palindromic sequence of motif 1 (PczcD-2), or the whole (PczcD-4) or half (PczcD-3) of the palindromic sequence, led to abolishment of the SczA-dependent activation in the presence of Zn2+, whereas a promoter fragment truncated just upstream of motif 1 (PczcD-2b) still showed SczA- and Zn2+-dependent activation (Fig. 4). The same was seen when Co2+ was used to induce expression (data not shown).
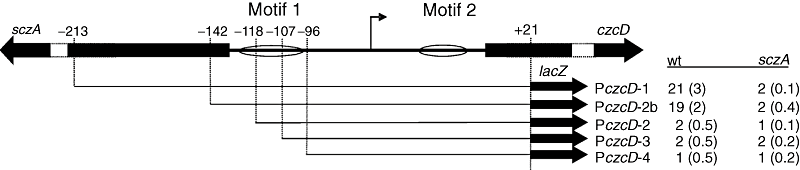
Subcloning of PczcD. A schematic drawing of PczcD truncations is shown. Positions of the truncations relative to the ATG start codon of czcD are given. The promoter is indicated with a flag. Ovals indicate the position of the two motifs that might function as operator sequences for SczA (motif 1 and motif 2). The table on the right gives β-galactosidase activity (Miller Units) of the promoter truncations in D39 wild-type (wt, strains MP120–MP124) and the ΔsczA mutant (sczA, strains MP130–MP134) grown in GM17 + 0.1 mM Zn2+. Standard deviations of three independent measurements are given between brackets.
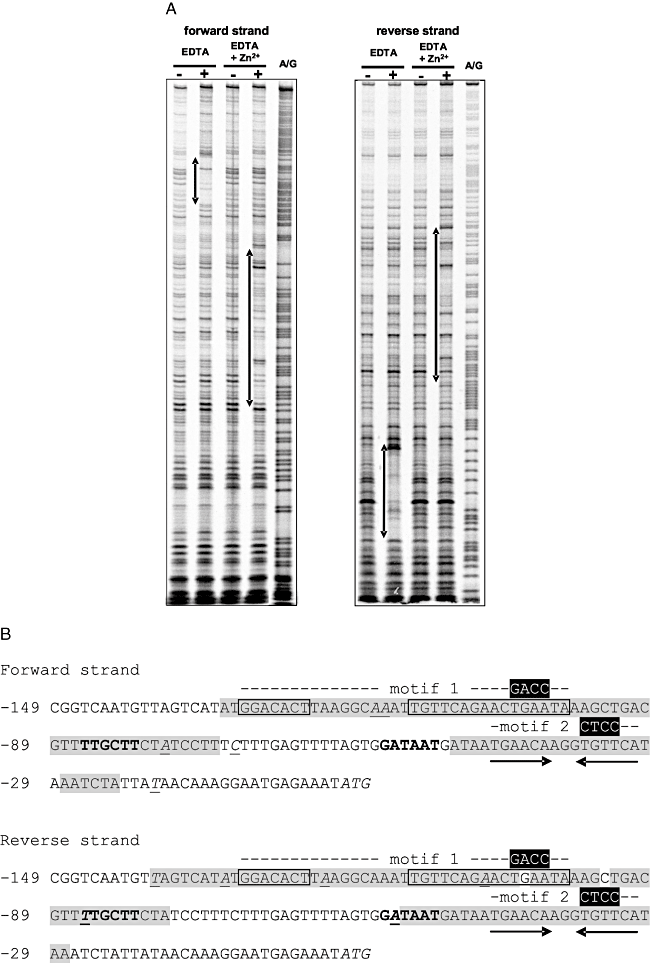
A. DNase I footprinting analyses of the binding of H6-SczA to PczcD. H6-SczA was added in concentrations of 0 M (–) or 7 × 10−8 M (+). A/G indicates lane with Maxam–Gilbert sequence ladder. EDTA and Zn2+ were added as indicated in a concentration of 0.3 mM. Left panel, the PczcD-1 PCR fragment was labelled with the forward primer PczcD-for1. Right panel, the PczcD-1 PCR fragment was labelled with the reverse primer PczcD-rev. Regions of protection are indicated with two-pointed arrows on the left of the lanes.B. Regions of protection in the PczcD sequence as found in the footprint analyses. The predicted SczA operator (motif 1) is boxed. The area protected in the footprint is in grey shading. Hypersensitive bases are in italics and underlined. The second SczA binding site (motif 2), a 15 bp inverted repeat, is indicated with arrows below the sequence. Putative −10 and −35 sequences are in bold. Base-pair mutations as introduced in PczcD-mut1 (and PsczA-mut1) and PczcD-mut2 are indicated in white above the sequence (positions −33 to −36). Numbers indicate base positions relative to the start codon (in italics) of czcD.
To determine whether motif 1 functions as a binding site for SczA, the promoter truncations were used in direct binding assays with purified H6-SczA (Fig. 5A), which was functional when expressed in S. pneumoniae (Fig. 2D). Surprisingly, H6-SczA specifically bound to all truncations, even to PczcD-4, which entirely lacks motif 1 (Fig. 5A). However, only with motif 1 present in its full length, including the bases upstream of the palindrome (PczcD-2b), binding resulted in complexes of higher molecular weight (MW) (Fig. 5A).
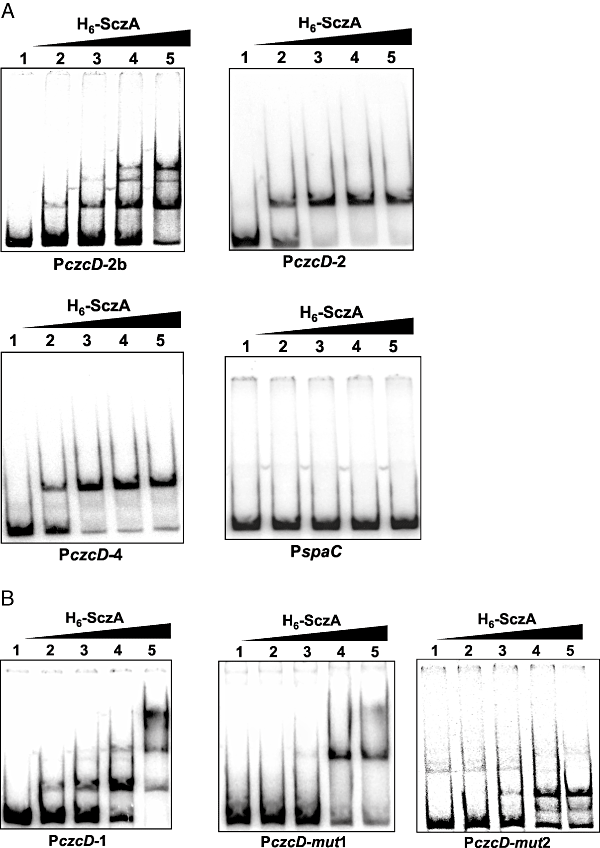
In vitro interaction of H6-SczA with PczcD.A. EMSAs of H6-SczA with PczcD truncations PczcD-2b, PczcD-2 and PczcD-4 (see Fig. 4). The spaC promoter was used as a negative control. Purified H6-SczA was added in concentrations of 0, 1.15 × 10−8, 2.3 × 10−8, 4.6 × 10−8 and 7 × 10−8 M in lanes 1–5 respectively.B. EMSAs of H6-SczA binding to PczcD-1 (see Fig. 4) and its derivatives PczcD-mut1 and PczcD-mut2 (see Fig. 8B). Conditions were the same as given under ‘A’.
Apparently, H6-SczA also binds to another, more downstream site in PczcD. Closer examination of this 3′ region of PczcD revealed a perfect inverted repeat (motif 2, 4, 8), of which the second half (5′-TGTTCA-3′) is identical to the first half of the palindromic part of the SczA operator (Fig. 8B), which therefore could function as an additional SczA binding site. This motif 2 is, however, not conserved in the other streptococci. To investigate the effect of motif 2 on the SczA-dependent activation of PczcD, four subsequent point-mutations were introduced in promoter fragment PczcD-1 (Fig. 8B). Expression from this PczcD fragment (PczcD-mut1) in the presence of Zn2+ or Co2+ was four times higher than PczcD-1, and it was also slightly expressed in GM17 (Fig. 6A). Conversely, introduction of four subsequent point-mutations in motif 1 (Fig. 8B, PczcD-mut2) abolished expression in the presence of Zn2+ or Co2+ completely (data not shown). Binding of purified H6-SczA to PczcD-mut1 resulted only in the band of higher MW (Fig. 5A), whereas binding to PczcD-mut2 resulted only in the low-MW bands (Fig. 5B). Thus, motif 2 functions as a binding site for SczA, giving rise to the complex of low MW. This interaction is not necessary for the activation of PczcD by SczA, but moderates activation of czcD expression. Motif 1, on the other hand, mediates the SczA-dependent activation of the czcD promoter.
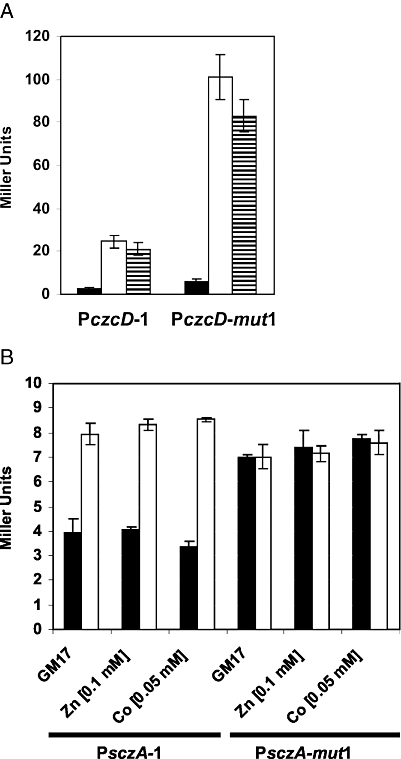
Effect of motif 2 on transcriptional regulation of the czcD and sczA promoters.A. Expression of PczcD-1–lacZ and PczcD-mut1–lacZ transcriptional fusions in D39 wild-type (strains MP120 and MP125) in GM17 (black bars), GM17 with 0.05 mM Co2+ (white bars) and GM17 with 0.1 mM Zn2+ (horizontally striped bars).B. Expression of PsczA-1–lacZ and PsczA-mut1–lacZ transcriptional fusions in D39 wild-type (strains MP127 and MP128, black bars) and ΔsczA (strains MP137 and MP138, white bars) in GM17 with the indicated concentrations of metal ions.
As SczA activates PczcD, we wondered whether it also affects expression of its own promoter, which is located on the same intergenic region (Fig. 1A), and whether this is mediated by motif 2. Therefore, expression of the wild-type (PsczA-1) and a sczA promoter with the four point-mutations in motif 2 (PsczA-mut1) was analysed in the wild-type strain and the sczA mutant (Fig. 6B). This showed that SczA has a twofold repressive effect on the expression of its own gene, which was, however, independent of the addition of the metal ions Zn2+ and Co2+ (Fig. 6B). The autorepressive effect of SczA was fully relieved upon mutation of motif 2 (Fig. 6B). Taken together, our experiments demonstrate the functionality of a SczA operator sequence (motif 1) that mediates activation of PczcD, as well as a second sequence (motif 2) that mediates autorepression of sczA and furthermore weakens activation of PczcD.
Zn2+-dependent binding of SczA to PczcD
To further elucidate the mechanism of transcriptional activation by SczA, the effect of metal ions on binding of H6-SczA to PczcD was studied. To prevent interference with the binding of metal ions to H6-SczA, running- and gel buffers without EDTA were used. Under these conditions, H6-SczA binding to PczcD-1 resulted only in the higher-MW complex (binding to motif 1, Fig. 7A), as opposed to the formation of both high- and low-MW complexes with the standard TBE running buffer that contains 2 mM EDTA, which was used for the shifts in Fig. 5A. Binding of H6-SczA to PczcD-1 in the absence of EDTA was stimulated by Zn2+, Co2+ and Ni2+, but not by Mn2+ and apparently Mg2+, which is present in a 5 mM concentration in the binding buffer (Fig. 7A and B). Addition of EDTA to the reaction mixture, however, led to formation of the lower-MW complex (Fig. 7A and B). Titration of Zn2+ in the reactions with EDTA indeed led to formation of the higher-MW complex again, while Mn2+ did not (Fig. 7A). Also Co2+ and Ni2+ led to formation of the higher band (Fig. 7B). The fact that the low-MW band was only seen upon addition of EDTA, suggests that the purified H6-SczA contains a low amount of metal ions, possibly Ni2+ as a consequence of the purification procedure, which favours formation of the higher-MW complex.
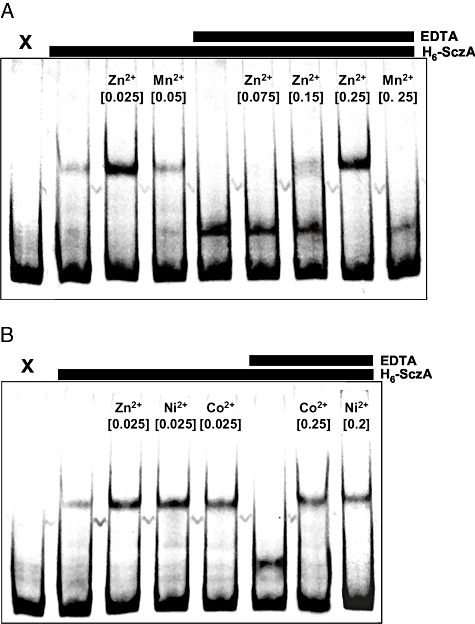
Modulation of binding of H6-SczA to PczcD by Zn2+, Ni2+ and Co2+ analysed with EMSAs.A. Effect of Zn2+ on binding of H6-SczA to PczcD-1. H6-SczA and EDTA were added as indicated above the gel in a concentration of 3.5 × 10−8 M and 0.3 mM respectively. Metal ions were added as indicated above the lanes, with concentrations in mM between brackets. The electrophoresis and gel buffer did not contain EDTA. X, free probe PczcD-1.B. Effect of Co2+ and Ni2+ on binding of H6-SczA to PczcD-1. Conditions were the same as described for A, except for the metal ions that are indicated in the figure.
To analyse the effects observed above in more detail, DNase I footprint analyses were performed with H6-SczA and PczcD, in the presence of EDTA and in the presence of both EDTA and Zn2+(Fig. 8A and B). In the presence of EDTA, only protection was seen of motif 2, whereas the addition of Zn2+ led to disappearance of this protected region and the appearance of a protected area comprising motif 1 (Fig. 8A and B). Thus, the data argue for a model in which binding of SczA to motif 1 in PczcD is stimulated by Zn2+, while SczA binding to motif 2 is relieved by Zn2+, in this way accomplishing the Zn2+-dependent activation of czcD expression (Fig. 9).
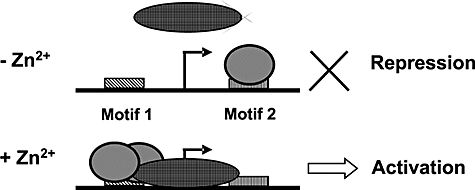
Schematic model of the mechanism of activation of PczcD by SczA depending on Zn2+. In the absence of Zn2+, SczA binds to motif 2, thereby blocking transcription from PczcD. In the presence of Zn2+, SczA binds to motif 1, which leads to activation of PczcD. Arrows indicate the czcD promoter. Big oval, RNA polymerase. Small oval, SczA. The binding of SczA to motif 1 in a higher oligomeric complex is indicated by the two overlapping SczA symbols, as opposed to the single (lower oligomeric state) SczA symbol bound to motif 2.
Discussion
In this study, we investigated the regulation of czcD, which we demonstrate to encode an important Zn2+-resistance determinant in the human pathogen S. pneumoniae. A novel TetR-family regulator, SczA, was identified that activates expression of czcD in the presence of Zn2+, Co2+ and Ni2+. Moreover, we identified and demonstrated the functionality of an operator sequence for SczA. TetR-family regulators are known to be involved in a variety of functions, such as regulation of antibiotic biosynthesis, osmotic stress and multidrug resistance, and they usually function as repressors (Ramos et al., 2005). To our knowledge, this is the first metalloregulatory protein belonging to the TetR family identified so far. In addition, it is one of a few examples of regulators from the TetR family that function as a transcriptional activator (Lin et al., 2000; Croxatto et al., 2002; Ramos et al., 2005; Novakova et al., 2005; Christen et al., 2006).
The CDF family, to which CzcD belongs, is an ubiquitous family of transporters present in prokaryotes and eukaryotes, which play a role in metal ion homeostasis, but also appear to fulfil other functions in the cell (Haney et al., 2005). We show that CzcD protects significantly against Zn2+ stress in S. pneumoniae, and to a lesser extent against Co2+ stress. Alignment shows that S. pneumoniae CczD contains the conserved aspartyl residues that are known to contribute to a metal ion binding site and metal ion transport in the CDF-family Zn2+ efflux pump YiiP of Escherichia coli (Wei and Fu, 2006). This suggests that CzcD also functions as a Zn2+ efflux pump in S. pneumoniae.
The observation that czcD in S. pneumoniae seems to be primarily involved in resistance towards Zn2+ raises the question as to the reason of the strong transcriptional response of PczcD to Co2+ and Ni2+. Possibly, there are other efflux pumps in S. pneumoniae that are involved in resistance against Co2+ and Ni2+. This would agree with the observed aberrant regulation of PczcD in the czcD mutant in response to Co2+ and Ni2+, but not to Zn2+. Other candidate heavy-metal-resistance genes encoded by the S. pneumoniae genome are spr1466 (cadD), a putative cadmium-resistance transporter, and spr1411, encoding a CDF-family protein. A possible third metal ion-resistance gene might be spr0641, which shows homology to the PerR-regulated gene pmtA in S. pyogenes involved in resistance to Zn2+ (Brenot et al., 2007).
The transcriptional response of czcD in S. pneumoniae shows similarity to that found in B. subtilis, where czcD is induced in the presence of Zn2+, Ni2+, Co2+ and Cu2+. However, the regulatory mechanism underlying this response is completely different, as in B. subtilis, an ArsR-type regulator, CzrA, functions as a repressor of czcD (Moore et al., 2005). Close homologues of CzrA (> 45% identity) are present in several Staphylococcus, Bacillus and Listeriae species, whereas SczA is only present (> 45% identity) in a small group of related streptococci, where the sczA gene is linked to czcD. Several other important groups of Gram-positive bacteria, such as lactococci and enterococci, do not seem to contain either regulator type, indicating a yet different regulatory factor governing expression of their czcD orthologues.
The microarray results show that SczA also regulates expression of spr1671, encoding a MerR-family regulator with homology (30% identity) to copper-responsive regulators, and adhB, which are located immediately downstream of czcD. The fact that adhB is regulated by SczA makes sense, as it is predicted to encode a Zn2+-containing alcohol dehydrogenase. AdhB was found in an STM screen to be important for virulence in a lung infection model (Hava and Camilli, 2002). Also, spr1671 (SP1856) seems important for virulence based on STM screens (Hava and Camilli, 2002; Hava et al., 2003). Thus, SczA could indirectly influence S. pneumoniae virulence via adhB and spr1671.
The functionality of a putative SczA operator, predicted on the bases of sequence analysis of PczcD regions of different streptococci, was confirmed by promoter subcloning, EMSAs and DNase I footprinting. The operator appears to consist of a palindromic sequence, as well as a stretch of seven bases located just upstream of the palindrome, together making up motif 1. In addition, a second binding site for H6-SczA was identified, that moderates expression of PczcD and is responsible for the autorepression by SczA. Binding of H6-SczA to motif 1 was increased by Zn2+, whereas binding to motif 2 was decreased in the presence of Zn2+. This indicates that in the absence of Zn2+, SczA binds to motif 2, thereby tightly shutting down PczcD. However, in the presence of Zn2+, SczA binds to motif 1, resulting in activation of PczcD. As repression of transcription from the czcD and sczA promoters mediated by motif 2 was not relieved in the presence of Zn2+, the relevance of SczA binding to motif 2 remains somewhat obscure. Maybe, this interaction is only destroyed under extreme metal ion intoxication, thereby giving an extra boost to czcD expression and, as a consequence, metal ion resistance. The observation that binding of H6-SczA to motif 1 resulted in a band of higher MW than binding of H6-SczA to motif 2, might indicate that monomeric SczA binds to motif 2, whereas an oligomeric conformation of SczA binds to motif 1 (Fig. 9).
Zn2+ has a profound influence on the immune function of the human body (Milanino et al., 1993; Shankar and Prasad, 1998; Schaible and Kaufmann, 2004; Thurnham et al., 2005). Generally, low Zn2+ levels lead to decreased functioning of the immune system, while physiologically normal concentrations [12–16 μM in plasma (Versieck, 1985)] secure normal functioning of the immune system (Shankar and Prasad, 1998; Rink and Gabriel, 2000; Ibs and Rink, 2003). High dosages of Zn2+ (0.1 mM) may even activate certain cells of the immune system (Ibs and Rink, 2003). Moreover, Zn2+ levels in the human body are increased during inflammation (Milanino et al., 1993; Thurnham et al., 2005). Thus, it is well possible that S. pneumoniae encounters fluctuating levels of Zn2+ during infection of the human body. One of the strategies S. pneumoniae could employ to cope with this is by activation of the czcD system in the presence of an elevated Zn2+ level. Thus, Zn2+ might be an important signal during the complex interplay between S. pneumoniae and humans.
Experimental procedures
Bacterial strains, growth conditions, materials and general DNA techniques
Bacterial strains are listed in Table 4. Growth of bacteria and DNA manipulation techniques were performed essentially as described previously (Kloosterman et al., 2006a,b). Metals were used as the following salts: ZnSO4, CoCl2, NiSO4, MgCl2, MnCl2, CuSO4, FeCl2 and FeCl3. D39 chromosomal DNA was used as a template for PCR reactions. Primers are listed in Table 5.
| Strain/plasmid | Description | Reference or source |
|---|---|---|
| Strain | ||
| S. pneumoniae | ||
| D39 | Serotype 2 strain, cps2 | Avery et al. (1944), Lanie et al. (2007) lab. P. Hermans |
| D39nisRK | D39 ΔbgaA::nisRK; TrmpR | Kloosterman et al. (2006a) |
| MP100 | D39 ΔsczA; SpecR | This work |
| MP101 | D39nisRKΔsczA; SpecR | This work |
| MP102 | D39 ΔczcD | This work |
| MP103 | D39 ΔbgaA::PczcD–lacZ; TetR | This work |
| MP105 | D39 ΔbgaA::Pspr1671–lacZ; TetR | This work |
| MP106 | D39 ΔbgaA::Pspr0183–lacZ; TetR | This work |
| MP107 | MP100 ΔbgaA::PczcD–lacZ; TetR SpecR | This work |
| MP109 | MP100 ΔbgaA::Pspr1671–lacZ; TetR SpecR | This work |
| MP110 | MP100 ΔbgaA::Pspr0183–lacZ; TetR SpecR | This work |
| MP111 | MP102 ΔbgaA::PczcD–lacZ; TetR | This work |
| MP112 | MP103 with mariner insertion in sczA SpecR | This work |
| MP120 | D39 ΔbgaA::PczcD-1–lacZ; TetR | This work |
| MP121 | D39 ΔbgaA::PczcD-2–lacZ; TetR | This work |
| MP122 | D39 ΔbgaA::PczcD-2b–lacZ; TetR | This work |
| MP123 | D39 ΔbgaA::PczcD-3–lacZ; TetR | This work |
| MP124 | D39 ΔbgaA::PczcD-4–lacZ; TetR | This work |
| MP125 | D39 ΔbgaA::PczcD-mut1–lacZ; TetR | This work |
| MP126 | D39 ΔbgaA::PczcD-mut2–lacZ; TetR | This work |
| MP127 | D39 ΔbgaA::PsczA-1–lacZ; TetR | This work |
| MP128 | D39 ΔbgaA::PsczA-mut1–lacZ; TetR | This work |
| MP130 | MP100 ΔbgaA::PczcD-1–lacZ; TetR SpecR | This work |
| MP131 | MP100 ΔbgaA::PczcD-2–lacZ; TetR SpecR | This work |
| MP132 | MP100 ΔbgaA::PczcD-2b–lacZ; TetR SpecR | This work |
| MP133 | MP100 ΔbgaA::PczcD-3–lacZ; TetR SpecR | This work |
| MP134 | MP100 ΔbgaA::PczcD-4–lacZ; TetR SpecR | This work |
| MP135 | MP100 ΔbgaA::PczcD-mut1–lacZ; TetR SpecR | This work |
| MP126 | MP100 ΔbgaA::PczcD-mut2–lacZ; TetR SpecR | This work |
| MP137 | MP100 ΔbgaA::PsczA-1–lacZ; TetR SpecR | This work |
| MP138 | MP100 ΔbgaA::PsczA-mut1–lacZ; TetR SpecR | This work |
| L. lactis | ||
| NZ9000 | MG1363 ΔpepN::nisRK | Kuipers et al. (1998) |
| E. coli | ||
| EC1000 | KmR; MC1000 derivative carrying a single copy of the pWV01 repA gene in glgB | Leenhouts et al. (1996) |
| Plasmid | ||
| pR412-T7 | SpecR; derivative of pR412 (Martin et al., 2000) | Bijlsma et al. (2007) |
| pORI13 | EmR; ori+repA-; promoterless lacZ, for single-copy chromosomal lacZ fusions | Sanders et al. (1998) |
| pORI38 | SpecR; ori+repA-; deletion derivative of pWV01 | Leenhouts et al. (1996) |
| pNZ8048 | CmR; nisin-inducible PnisA | de Ruyter et al. (1996) |
| pNG8048E | CmR EmR; nisin-inducible PnisA, pNZ8048 derivative containing emR gene to facilitate cloning | Laboratory collection |
| pORI280 | EmR; ori+repA-; deletion derivative of pWV01; constitutive lacZ expression from P32 promoter | Leenhouts et al. (1996) |
| pPP2 | AmpR TetR; promoter-less lacZ. For replacement of bgaA (spr0565) with promoter–lacZ fusions. Derivative of pTP1 | Halfmann et al. (2007) |
| pMP1 | pORI280 ΔczcD | This work |
| pMP2 | pPP2 PczcD–lacZ | This work |
| pMP3 | pPP2 Pspr1671–lacZ | This work |
| pMP5 | pPP2 Pspr0183–lacZ | This work |
| pMP6 | pNG8048E carrying H6-sczA downstream of PnisA | This work |
| pMP7 | pPP2 PczcD-1–lacZ | This work |
| pMP8 | pPP2 PczcD-2–lacZ | This work |
| pMP9 | pPP2 PczcD-2b–lacZ | This work |
| pMP10 | pPP2 PczcD-3–lacZ | This work |
| pMP11 | pPP2 PczcD-4–lacZ | This work |
| pMP12 | pPP2 PczcD-mut1–lacZ | This work |
| pMP13 | pPP2 PsczA-1–lacZ | This work |
| pMP14 | pPP2 PsczA-mut1–lacZ | This work |
| pMP15 | pPP2 PczcD-mut2-lacZ | This work |
- TrmpR, trimethoprim resistance; SpecR, spectinomycin resistance; EmR, erythromycin resistance; TetR, tetracycline resistance; KmR, kanamycin resistance; CmR, chloramphenicol resistance.
| Name | Nucleotide sequence (5′ to 3′) (restriction enzyme sites underlined) | Restriction site |
|---|---|---|
| PczcD-1 | CGGAATTCTAGATGGCTTTTTTGGTTTTGCTG | EcoRI |
| PczcD-2 | CGGGATCCGCAGACTCAGAATAGACTCATTC | BamHI |
| PczcD_for1 | CGGAATTCCTCGTAGCCCTTAGCATTCA | EcoRI |
| PczcD_for2 | CGGAATTCCAAATTGTTCAGAACTGAAT | EcoRI |
| PczcD_2b | CGGAATTCTGTTAGTCATATGGACACTTAAGG | EcoRI |
| PczcD_for3 | CGGAATTCGAACTGAATAAAGCTGACG | EcoRI |
| PczcD_for4 | CGGAATTCAGCTGACGTTTTGCTTCTAT | EcoRI |
| PczcD_rev | CGGGATCCAACAGCATATTTTGCCTTCA | BamHI |
| PczcD-mut1.1 | GGAGCCTTGTTCATTATCATTATCCAC | – |
| PczcD-mut1.2 | ATAATGAACAAGGCTCCCATAAATCTATTATAACAAAGG | – |
| PczcD-mut2.1 | GACCTAAAGCTGACGTTTTGCTTC | – |
| PczcD-mut2.2 | CAAAACGTCAGCTTTAGGTCGTTCTGAACAATTTGCCTTAAGTG | – |
| czcD-KO-1 | TGCTCTAGAAGGTCAATGTCTCGATAAAG | XbaI |
| czcD-KO-2 | AGCATATTTTGCCTTCATATTTC | – |
| czcD-KO-3a | ATATGAAGGCAAAATATGCTAGTTATGAGCATCAACATTAGa | – |
| czcDKO-4 | GAAGATCTCTGTAGCTGAGACAAGCGC | BglII |
| PsczA-for1 | CGGGATCCCTCGTAGCCCTTAGCATTCA | BamHI |
| PsczA-rev | CGGAATTCAACAGCATATTTTGCCTTCA | EcoRI |
| Spr1673KO-1 | CTAACAGATTGATAGTAATCG | – |
| Spr1673KO-2b | TCCTCCTCACTATTTTGATTAGATACGGCGGTCAATGTTAGTCb | – |
| Spr1673KO-3b | CGTTTTAGCGTTTATTTCGTTTAGTTTATCTAGACCTTCTCATTCCb | – |
| Spr1673KO-4 | CTGGACGGCAAGGGCTGGAC | – |
| spr1671-1 | TGCTCTAGAAAGATTTTGCATCCGCAACC | XbaI |
| spr1671-2.1 | CGGGATCCCGGCAGATTTAATATTCACAC | BamHI |
| spr1671-2 | CGGGATCTCGGCAGATTTAATATTCACAC | – |
| spr1671-3 | CATGCCATGGGAGCGCTTGTCTCAGCTACA | – |
| spr1671-4 | GAAGATCTCAAAGCCCATACGCCCTCC | – |
| TMr_1 | TGCATTTAATACTAGCGACGCCATCTATGTGTC | – |
| TMr_4 | GGATCCATTCGCGTCAATTCGAGGGG | – |
| Spec_Fp | CTAATCAAAATAGTGAGGAGG | – |
| Spec_Rp | ACTAAACGAAATAAACGC | – |
| PspaC-1 | CCAGTCCAGACTATTCGG | – |
| PspaC-2 | CAGAGGTTGTTCTGG | – |
| Pspr183-1.2 | CGGAATTCCAACCTAAGGTGATTGTGG | EcoRI |
| Pspr183-2.2 | CGGGATCCGAATTTCTGTAATAATTCGC | BamHI |
| tetR-OX-1-H6 | CGAGCCATCATGACTCATCATCATCATCATCATAACATTGACCG | RcaI |
| tetR-OX-2 | CGGGATCCTCAATTTTTAGGAATGAGAAG | BamHI |
- a. Overlap with czcD-KO-2 in italics.
- b. Overlap with specR gene in bold.
Random mutagenesis screen using the mariner transposon
Chromosomal DNA of S. pneumoniae R6 was mutagenized with the Himar1 MarC9 transposase essentially as described (Lampe et al., 1996; Martin et al., 2000). pR412-T7 (Bijlsma et al., 2007), a derivative of pR412 (Martin et al., 2000), containing an outward-facing T7 promoter on each side of the mini-transposon, was used as the source of the 1146 bp long spec mariner mini-transposon. Mutated R6 chromosomal DNA was transformed to strain R6 to generate a mutant library of approximately 20 000 colony-forming units. Chromosomal DNA of this library was isolated and used to transform strain D39 ΔbgaA::PczcD–lacZ (MP103), and was plated on GM17 with 1% sheep blood, 0.006% Xgal, 1.5 μg ml−1 tetracycline and 130 μg ml−1 spectinomycin, and 0.25 mM ZnSO4. Plates were incubated for 2 days at 37°C. Sites of transposon insertion were determined using an inverse PCR approach. In short, chromosomal DNA of mutants was digested with AvaII and self-ligated at a concentration of 0.02 μg DNA per μl. PCR was performed on this ligation mixture with primers TMr_1 and TMr_4, and the resulting PCR products were isolated from gel with the Qiagen gel-extraction kit and sequenced.
Construction of mutants
A mutant of sczA (spr1673) was made by allelic-replacement mutagenesis. In short, primers Spr1673KO-1/Spr1673KO-2 and Spr1673KO-3/Spr1673KO-4 were used to generate PCR products of the approximately 500 bp left and right flanking regions of sczA, which were, by means of overlap extension PCR (Song et al., 2005), fused to a spectinomycin-resistance gene amplified with primers Spec_Fp and Spec_Rp from plasmid pORI38. The resulting PCR product was transformed to D39 and D39nisRK, yielding strains MP100 and MP101, and the mutation was verified by PCR and sequencing.
An in-frame deletion of czcD (spr1672) was constructed using pORI280 (Leenhouts et al., 1996; Kloosterman et al., 2006a). In short, primer pairs czcD-KO-1/czcD-KO-2 and czcD-KO-3/czcD-KO-4 were used to generate approximately 600 bp PCR fragments of the left and right flanking regions of czcD, which were fused in a separate PCR reaction and cloned into the XbaI/BglII sites of pORI280 in E. coli EC1000, yielding pMP1. A czcD mutant in D39 (MP102) was obtained following the procedure described before (Kloosterman et al., 2006a), and the mutation was verified by PCR and sequencing.
Construction of lacZ fusions
Transcriptional fusions to lacZ were constructed in plasmid pPP2. Primer pairs PczcD-1/PczcD-2, spr1671-1/spr1671-2.1 and Pspr183-1.2/Pspr183-2.2 were used to generate PCR fragments spanning the sczA–czcD, czcD–spr1671 intergenic regions and the nrdD promoter respectively, and cloned in the EcoRI/BamHI, XbaI/BamHI and EcoRI/BamHI sites of pPP2, using E. coli EC1000 as a host, yielding plasmids pMP2–pMP5. Correct constructs were transformed to D39 wild-type and MP100 to generate ectopic lacZ fusions in the bgaA locus, yielding strains MP103, MP105, MP106 and MP107, MP109 and MP110 respectively. The PczcD–lacZ fusion was introduced in the czcD mutant (MP102) as well, giving strain MP111.
Subcloning and mutation of the czcD and sczA promoters
Subclones (PczcD-1, PczcD-2, PczcD-2b, PczcD-3 and PczcD-4) of the PczcD were inserted in the EcoRI/BamHI sites of the lacZ reporter plasmid pPP2, using primer pairs PczcD_for1/PczcD_rev, PczcD_for2/PczcD_rev, PczcD_2b/PczcD_rev, PczcD_for3/PczcD_rev and PczcD_for4/PczcD_rev, giving plasmids pMP7 to pMP11. Four subsequent point-mutations were introduced into motif 2 in the czcD promoter as follows. PCR products were generated on D39 chromosomal DNA with primers PczcD-mut1.1/PczcD_for-1 and PczcD-mut1.2/PczcD_rev. These PCR products overlap for 17 bases and were fused in a separate PCR reaction with the outer primers PczcD-mut1.1 and PczcD_rev. The resulting PCR product was cloned as an EcoRI/BamHI fragment into pPP2, yielding plasmid pMP12. In the same way, four subsequent point-mutations were introduced in motif 2 in the czcD promoter, using primers PczcD-mut2.1/PczcD_for-1 and PczcD-mut2.2/PczcD_rev, yielding plasmid pMP15. The cloned sequences of these plasmids were verified by DNA sequencing, and they were subsequently transformed to D39 wild-type and its isogenic sczA mutant, giving strains MP120–MP126 and MP130–MP136.
The wild-type (PsczA-1) and mutated (PsczA-mut1) sczA promoters were amplified with primers PsczA-for1 and PsczA-rev from plasmids pMP7 and pMP12 respectively, and cloned as EcoRI/BamHI fragments into pPP2. The resulting constructs were verified by DNA sequencing and introduced into strains D39 and MP100, yielding strains MP127, MP128, MP137 and MP138.
Construction of an SczA overexpression construct and purification of H6-SczA
A his-tagged variant of sczA (H6-sczA) was PCR amplified with primers tetR-OX-1-H6/tetR-OX-2, digested with RcaI and BamHI and cloned in the NcoI/BamHI sites of pNG8048E, using Lactococcus lactis NZ9000 as the cloning host, giving plasmid pMP6.
Overexpression in L. lactis and purification of H6-SczA was performed essentially as described (Kloosterman et al., 2006b). H6-SczA was eluted from the Ni-NTA (Qiagen) beads with 250 mM imidazole. The protein was checked with SDS-PAGE and stored as frozen aliquots with 10% glycerol at −80°C.
Microarray analyses
For DNA microarray analysis, D39 wild-type and its isogenic sczA mutant (MP100) were grown as four biological replicates in GM17 with 0.05 mM CoCl2 and harvested at an OD595 of approximately 0.3. All other procedures regarding microarray analyses were performed as described before (Kloosterman et al., 2006b). Statistical analysis was performed as described (van Hijum et al., 2005). A gene was considered differentially expressed when the Bayesian P-value < 0.001 and a false discovery rate < 0.01, and when at least five measurements were available. The raw and processed data are available at http://molgen.biol.rug.nl/publication/scza_data.
Growth experiments
Growth experiments were performed in microtiterplates in 220 μl volumes. As the inoculum, aliquots of cells frozen in the mid-exponential phase were used, washed once with the proper medium and diluted 1 to 40 for inoculation.
β-Galactosidase assays
β-Galactosidase assays were performed essentially as described (Kloosterman et al., 2006b). Strains were harvested at mid-logarithmic phase of growth. All experiments were performed at least in triplo.
SczA–DNA interaction studies
Electrophoretic mobility shift assays (EMSAs) were performed essentially as described previously (den Hengst et al., 2005). PCR products of PczcD (PczcD-1, PczcD-2, PczcD-2b, PczcD-4 and PczcD-mut1 and PczcD-mut2) were made with the primer pairs as given under ‘subcloning of PczcD’. As a negative control, a PCR fragment of the PspaC promoter was amplified with primers Pspac-1/Pspac-2 from plasmid pDG148 (Stragier et al., 1988). The binding buffer was composed of 20 mM TrisHCl, pH 8.0, 5 mM MgCl2, 0.1 mM dithiotreitol, 8.7% (w/v) glycerol, 62.5 mM KCl, 25 μg ml−1 bovine serum albumin, 50 μg ml−1 poly(dI–dC) and 3000 cpm of either [γ-32P]ATP-labelled or [γ-33P]ATP-labelled PCR product. Purified H6-SczA, EDTA and metal ions were added as specified in the Results section. Reactions (20 μl) were incubated for 15 min at 37°C, after which they were run on a 6% poly-acrylamide gel for 80 min at 90 V. DNase I footprinting was performed essentially as described (den Hengst et al., 2005). In total, 150 000 cpm of [γ-33P]ATP-labelled PCR product PczcD-1, amplified either with [γ-33P]-labelled primer PczcD-for1 (forward strand) or with [γ-33P]-labelled primer PczcD-rev (reverse strand), was used as probe in 40 μl of binding buffer containing H6-SczA, EDTA and Zn2+ as specified in the Results. Buffer and reaction conditions were the same as for the EMSAs.
Reverse transcription (RT)-PCR
Reverse transcription reactions were performed as described under Microarray analysis on total RNA isolated from S. pneumoniae D39 grown in GM17 + 0.05 mM Co2+, except that amino-allyl dUTP was replaced by dTTP. In parallel, reactions were performed in the same way, except that the reverse transcriptase enzyme Superscript III was omitted. These reactions were used as negative controls. PCRs were performed on 1/100 part of the RT reactions with primers as specified in the Results. As a positive control, PCRs were performed on 10 ng per reaction of chromosomal DNA of D39.
Acknowledgements
We thank Peter Burghout (UMC St Radboud, Nijmegen, the Netherlands) for the gift of chromosomal DNA mutagenized with the mariner transposon. We thank Robert Witwicki for skilful technical assistance. We thank Don Morrison (University of Illinois, Chicago, USA) for the generous gift of CSP-1. This work was supported by IOP Grant IGE3002 of the Dutch Ministry of Economic Affairs.




