Mutations in Mycobacterium tuberculosis Rv0444c, the gene encoding anti-SigK, explain high level expression of MPB70 and MPB83 in Mycobacterium bovis
Summary
It has recently been advanced that Mycobacterium tuberculosis sigma factor K (SigK) positively regulates expression of the antigenic proteins MPB70 and MPB83. As expression of these proteins differs between M. tuberculosis (low) and Mycobacterium bovis (high), this study set out to determine whether M. bovis lacks a functional SigK repressor (anti-SigK). By comparing genes near sigK in M. tuberculosis H37Rv and M. bovis AF2122/97, we observed that Rv0444c, annotated as unknown function, had variable sequence in M. bovis. Analysis of in vitro mpt70/mpt83 expression and Rv0444c sequencing across M. tuberculosis complex (MTC) members revealed that high-level expression was associated with a mutated Rv0444c. Complementation of M. bovis bacillus Calmette-Guerin Russia, a high producer of MPB70/MPB83, with wild-type Rv0444c resulted in a significant decrease in mpb70/mpb83 expression. Conversely, a M. tuberculosis H37Rv mutant which expressed sigK but not Rv0444c manifested the M. bovis phenotype of high-level MPB70/MPB83 expression. Further support that Rv0444c encodes the anti-SigK was obtained by yeast two-hybrid studies, where the N-terminal region of Rv0444c-encoded protein interacted with SigK. Together these findings indicate that Rv0444c encodes the regulator of SigK (RskA) and mutations in this gene explain high-level MPT70/MPT83 expression by certain MTC members.
Introduction
The Mycobacterium tuberculosis complex (MTC) comprises pathogenic organisms for humans (M. tuberculosis sensu stricto), cattle (Mycobacterium bovis) and a number of other hosts, including seals, voles and goats (Mycobacterium pinnipedii, Mycobacterium microti and Mycobacterium caprae respectively) (Mostowy et al., 2005; Smith et al., 2006). Post-genomic study of these naturally occurring variants of the tubercle bacillus has revealed limited genetic variability, with 99.9% similarity at the nucleotide level and about 2% variability in genome content through insertion/deletion events (Sreevatsan et al., 1997; Fleischmann et al., 2002; Gutacker et al., 2002; Garnier et al., 2003). Nonetheless, an important difference between these closely related organisms is observed in their antigenic protein repertoire. For instance, both M. microti and the dassie bacillus are natural mutants of the RD1-region encoding ESAT-6 (Pym et al., 2002; Mostowy et al., 2004a), a region also implicated in the attenuation of bacillus Calmette–Guerin (BCG) vaccine strains (Pym et al., 2002; Lewis et al., 2003). Furthermore, sequence comparison of M. bovis AF2122/97 to M. tuberculosis H37Rv has revealed a number of differences in the coding sequences of antigenic PE/PPE PGRS proteins (Garnier et al., 2003).
A long-recognized and striking phenotypic difference between M. bovis and M. tuberculosis is in the production of antigenic proteins MPB70 and MPB83. M. bovis produces these proteins at high levels in vitro, whereas, for M. tuberculosis, there is a very low in vitro production of the corresponding proteins, MPT70 and MPT83 (by convention, ‘mpt ’ refers to a gene in M. tuberculosis while ‘mpb’ refers to the corresponding gene in M. bovis) (Hewinson et al., 1996). Further studies demonstrated that with the exception of M. bovis, which had elevated expression of MPB70, all MTC members tested, including M. tuberculosis, Mycobacterium africanum and M. microti, were low producers of this antigen (Liebana et al., 1996; Cousins et al., 2003). Interestingly, production of these proteins by M. bovis BCG strains is also variable (Wiker et al., 1996). BCG strains obtained from the Pasteur Institute before 1927 (‘early strains’) produce these proteins at high levels unlike strains obtained in 1931 or later (‘late strains’) where production is low (Charlet et al., 2005). Recently our laboratory has demonstrated that a mutation in sigK, the gene encoding the extracytoplasmic function (ECF) sigma factor K, explains the difference in MPB70/MPB83 expression between the two groups of BCG strains (Charlet et al., 2005). Given that SigK is a positive regulator of MPB70 and MPB83, it was reasoned that a second polymorphism in SigK-mediated control of MPB70/MPB83 expression might explain their differential production between M. tuberculosis and M. bovis.
Most ECF sigma factors and their cognate anti-sigma genes are adjacent to each other and are co-transcribed (Missiakas and Raina, 1998; Bashyam and Hasnain, 2004). The repressor of the ECF sigma factor (the anti-sigma) binds to it and inhibits its activity. Upon an extracellular stimulus, the repressor dissociates from the sigma factor, which now becomes available and binds to the promoters of the target genes to initiate their transcription (Missiakas and Raina, 1998; Bashyam and Hasnain, 2004). As SigK is implicated in the regulation of MPB70/MPB83 expression, we hypothesized that the phenotypic difference between M. tuberculosis and M. bovis may be due to a dysfunctional anti-SigK in M. bovis. To test this, we employed the recently derived phylogenetic scenario for MTC evolution to search candidate genes for polymorphisms that correlated with the expression phenotype, and then performed molecular studies to document that Rv0444c encodes the anti-SigK, and that mutations in this gene explain the variable expression of MPB70 and MPB83 across different MTC members.
Results
Differential expression of MPT70 and MPT83 expression across MTC members
Differential in vitro expression of MPB70 has been described in M. tuberculosis and M. bovis as well as other traditional MTC members (Liebana et al., 1996; Cousins et al., 2003). Using the revised MTC phylogeny, we documented MPT70 and MPT83 expression across a panel of MTC members to specifically determine which organisms manifest altered expression. By quantitative reverse transcription-PCR (qRT-PCR) coupled with a molecular beacon to analyse mRNA levels of mpt70, our results demonstrate that this gene is differentially expressed across MTC members in vitro. Specifically, M. tuberculosis H37Rv and H37Ra, M. africanum and M. microti had low expression whereas M. caprae, M. bovis, BCG Russia and the Oryx bacillus were high expressors of mpt70 (Fig. 1A). To show that differential expression of mpt70 (Fig. 1A) also affects protein levels, we performed a Western blot analysis on culture filtrates using a rabbit polyclonal antibody against MPB70. As shown in Fig. 1B, the results correlate perfectly with the qRT-PCR data. We previously noted that the expression of mpb70 by BCG strains is coupled to mpb83 expression in vitro (Charlet et al., 2005). To determine whether this is also the case across natural MTC members, we tested MPT83 expression by Western blot using a polyclonal rabbit antibody; the results parallel those of MPT70 (Fig. 1C). A polyclonal antibody raised against MPB64, a secreted antigenic protein whose expression is independent of SigK, was used as a loading control. As evidenced in Fig. 1D, all strains produced this protein. In addition, for each member of the MTC, we also compared the in vitro transcriptome against that of M. tuberculosis H37Rv, to screen for important transcriptional differences across organisms (data provided in Table S1). Microarray-based interrogation revealed that upregulation of mpb70, mpb83 and neighbouring genes (dipZ, Rv2876) by M. caprae, M. bovis, BCG Russia and the Oryx bacillus figured prominently among their principal differences in expression compared with M. tuberculosis. Based on these results, MTC members analysed could be divided into two groups: M. tuberculosis H37Rv and H37Ra, M. africanum and M. microti had low in vitro production of MPT70 and MPT83, while high production was observed for M. bovis, BCG Russia (early BCG), M. caprae and the Oryx bacillus.
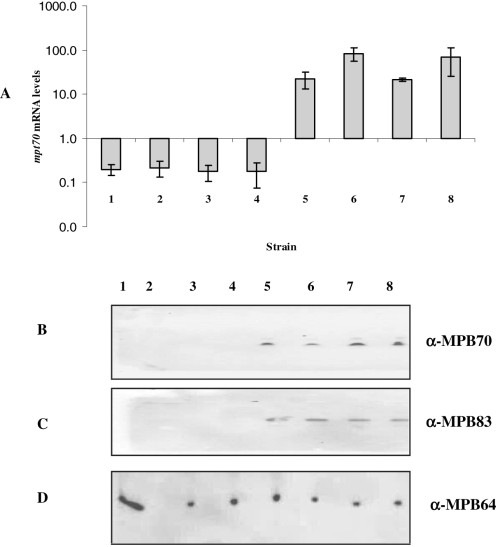
A. In vitro mpt70 mRNA levels across different MTC members. mRNA levels were measured by molecular beacon RT-PCR. Values represent the average of at least two independent RT-PCR reactions and are normalized to sigA mRNA.B–D. Western blot analysis of MPT70 (B), MPT83 (C) and MPT64 (D) production across MTC members. MPT64, a secreted antigen not regulated by SigK, was used as loading control. (B) and (D) are SDS-PAGE of culture filtrate proteins probed with a rabbit polyclonal antibody against MPB70 and a mouse polyclonal antibody against MPB64 respectively. (C) represents SDS-PAGE of cell extracts immunoblotted with a rabbit polyclonal antibody against MPB83. MTC members used are as follows: 1, M. tuberculosis H37Rv; 2, M. tuberculosis H37Ra; 3, M. africanum; 4, M. microti; 5, Oryx bacillus; 6, M. caprae; 7, M. bovis; 8, M. bovis BCG Russia.
Expression of mpt70 and mpt83 in M. tuberculosis is induced inside THP-1 macrophages
The low levels of MPT70 and MPT83 in M. tuberculosis have been documented in vitro (Hewinson et al., 1996). However, during intracellular infection, the organism modulates its expression, and mpt70/mpt83 figure among the genes reported to be strongly induced during adaptation to the phagosomal environment (Schnappinger et al., 2003). In this report, confirmation of microarray results by qRT-PCR was only performed for mpt83. To confirm that mpt70 expression is also inducible in M. tuberculosis, THP-1 macrophages were infected with M. tuberculosis H37Ra and virulent H37Rv, and bacterial mRNA was isolated to study mpt70 expression by qRT-PCR. As shown in Fig. 2, mpt70 levels are significantly higher in both H37Ra and H37Rv after 24 h inside THP-1 cells pretreated with interferon-γ (IFN-γ) as compared with non-stimulated cells, indicating that mpt70 is induced in stimulated macrophages (Fig. 2). Similar results were obtained for mpb83 (data not shown). These data imply that the wild-type expression phenotype, as exemplified by M. tuberculosis, is low in vitro and inducible during infection.
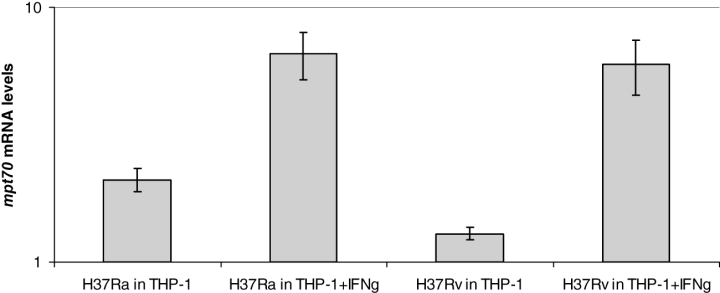
Induction of mpt70 expression in M. tuberculosis H37Rv and H37Ra inside THP-1 cells. mRNA levels were measured by molecular beacon RT-PCR. Data are expressed as the ratio between mRNA levels inside THP-1 versus mRNA levels of the same organism during extracellular growth in vitro. Values represent the average of at least two independent RT-PCR reactions and are normalized to sigA mRNA.
Genetic basis behind MPT70/MPT83 differential expression
To elucidate the genetic reason for the variable expression of MPT70 and MPT83 across MTC members, we first focused our attention on M. bovis (high producer) and M. tuberculosis H37Rv (low producer), whose whole genome sequences are publicly available (http://genolist.pasteur.fr/TubercuList, http://genolist.pasteur.fr/BoviList). The sequences of M. bovis mpb70 and mpb83 are identical to their M. tuberculosis counterparts mpt70 and mpt83. As both organisms also have the identical sequence of sigK, the gene encoding the positive regulator of the two antigens (Charlet et al., 2005), we hypothesized that M. bovis may lack a functional anti-SigK. By analysing the sequence of the genes in the vicinity of sigK in M. tuberculosis H37Rv versus M. bovis 2122, we found that only one gene, Rv0444c, located immediately downstream of sigK, varies between the two organisms. In M. bovis, a high producer of MPB70 and MPB83, Rv0444c harbours two non-synonymous single nucleotide polymorphisms (SNPs): C320T and C551T, resulting in an amino acid change of glycine to aspartic acid and glycine to glutamic acid respectively. We then sequenced Rv0444c across different MTC members and found that members with low-level production of MPT70/MPT83 have the wild-type gene (H37Rv sequence) whereas members with high-level expression of these antigens have a mutated Rv0444c (Table 1). Of note, the Oryx bacillus, which produces MPT70/MPT83 at high levels in vitro (Fig. 1), has a different Rv0444c mutation than M. bovis; the stop codon has been substituted with a serine, resulting in a read-through mutation that may affect protein translation and/or stability. The observation that the gene immediately neighbouring sigK had suffered two independent mutations that were associated with the same expression phenotype strongly implicated Rv0444c as the gene encoding anti-SigK.
| MTC member | NT@320 | NT@551 | NT@698 | MPT70/MPT83 production |
|---|---|---|---|---|
| M. tuberculosis H37Rv | C | C | G | Low |
| M. tuberculosis H37Ra | C | C | G | Low |
| M. africanum | C | C | G | Low |
| M. microti | C | C | G | Low |
| Oryx bacillus | C | C | C | High |
| M. caprae | T | T | G | High |
| M. bovis | T | T | G | High |
| Early BCG (Russia) | T | T | G | High |
- The changes from C to T result in G320D and G551E in M. caprae M. bovis, and BCG Russia. Changes from G to C in the Oryx bacillus results in stop codon replaced by Serine. SNPs in Rv0444c correlate with high mpb70 expression.
Complementation of BCG Russia with wild-type Rv0444c results in a significant decrease in mpb70/83 expression
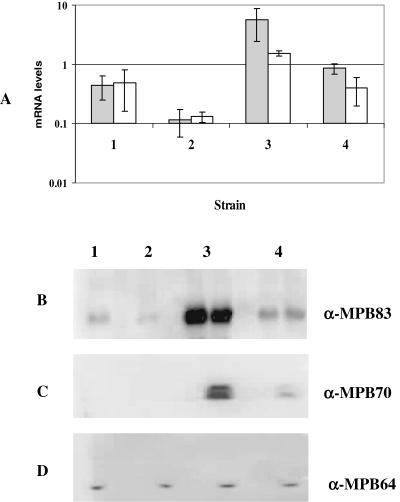
To functionally prove that Rv0444c gene product is the anti-SigK, we complemented BCG Russia, which has a mutated Rv0444c and is a high producer of MPB70/83, with the wild-type Rv0444c amplified from M. tuberculosis H37Rv, expressed under the mycobacterial optimal promoter (MOP) which consists of the BCGhsp70 promoter and the Escherichia coli tac promoter (George et al., 1995). To control for gene copy, we separately complemented BCG Russia with an overexpressed mutant Rv0444c (amplified from BCG Russia itself). Overexpression of wild-type and mutant Rv0444c was verified by qRT-PCR with SYBR green (data not shown). Next, using qRT-PCR, coupled with molecular beacons, we observed that addition of wild-type Rv0444c resulted in a pronounced decrease in mpb70/83 expression as compared with BCG Russia complemented with the empty vector, but complementation with the mutant Rv0444c had no effect in mpb70/83 expression (Fig. 3). This indicated that the decrease in expression we observed was specifically due to the introduction of wild-type Rv0444c.
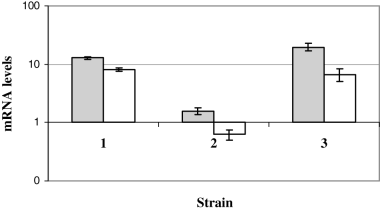
Expression of mpb70 and mpb83 in Russia complements. Values represent the average of at least two independent cDNA samples and are normalized to sigA mRNA. 1, Russia-PMH (Russia complemented with empty vector); 2, Russia::wtRv0444c (Russia complemented with wild-type Rv0444c amplified from M. tuberculosis H37Rv); 3, Russia::mutRv0444c (Russia complemented with mutant Rv0444c amplified from BCG Russia). Grey bars, mpb70; white bars, mpb83.
Lack of Rv0444c expression results in overexpression of mpt70 and mpt83
As a second means of proving that Rv0444c encodes the anti-SigK, we aimed to determine whether the lack of a functional Rv0444c in M. tuberculosis would result in an overexpression of mpt70/mpt83. To this end, we constructed a M. tuberculosis H37Rv mutant that expresses sigK but not Rv0444c in the following manner. A ΔsigK mutant was engineered by allelic exchange, replacing sigK with a kanamycin cassette using the pKO knock-out plasmid as described in Lewis et al. (2003). Deletion of sigK and its replacement by the kanamycin gene was confirmed by Southern blot analysis (Fig. 4A and B) and sequence analysis of the mutant indicated that the kanamycin cassette inserted between nucleotide positions 533 815 and 534 397 of the M. tuberculosis H37Rv genome resulting in the complete removal of sigK (Fig. 4C). As sigK and Rv0444c are in close proximity to each other (only 43 bp between them) and predicted to be co-transcribed, we analysed the expression of both sigK and Rv0444c in the deletion mutant by qRT-PCR. As expected, neither gene was expressed in the mutant, and hence the sigK deletion created a polar mutation affecting Rv0444c expression (Fig. 5A). Next, we separately complemented this strain with sigK alone (single gene complement) and with both sigK and Rv0444c gene (dyad complement). As shown in Fig. 5A, the single gene complement only expressed sigK whereas the dyad complement expressed both sigK and Rv0444c. These data were confirmed by a Western blot analysis of the Rv0444c gene product (Fig. 5B). We then analysed the expression of mpt70 and mpt83 in these clones by qRT-PCR. As shown in Fig. 6A, the single gene complement, which expressed sigK but not Rv0444c, expressed high levels of mpt70/mpt83 in vitro. The expression of mpt70/mpt83 in the ΔsigK::dyad strain was at low levels, similar to H37Rv (Fig. 6A). In perfect agreement with the qRT-PCR data, Western blot analysis of the same strains demonstrated that the sigK complement (ΔsigK::sigK) became a high producer of MPT70 and MPT83 proteins, whereas the dyad complement expressed MPT70 and MPT83 at levels similar to H37Rv (Fig. 6B and C). These results once again showed that Rv0444c encodes a protein with anti-SigK activity.
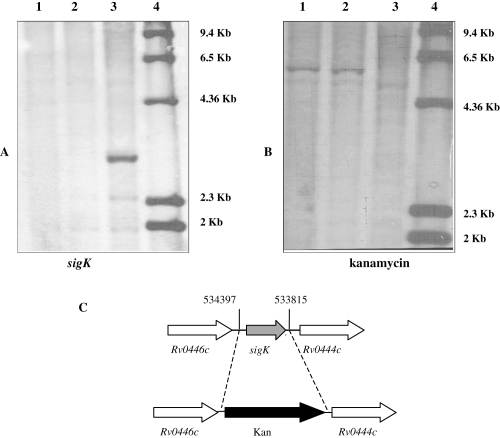
Confirmation of sigK deletion. Southern blot analysis of sigK (A) and kanamycin (B) in M. tuberculosis H37Rv and two independent clones of sigK deletion mutants. Genomic DNA was digested with PvuII, and probed with the specified probe. 1, ΔsigK clone 2-2; 2, ΔsigK clone 2-5; 3, H37Rv; 4, ladder with sizes of the bands marked on the right. (C) sigK locus and schematics of the kanamycin cassette insertion in the mutant. Numbers refer to the nucleotide position in the H37Rv genome sequence.
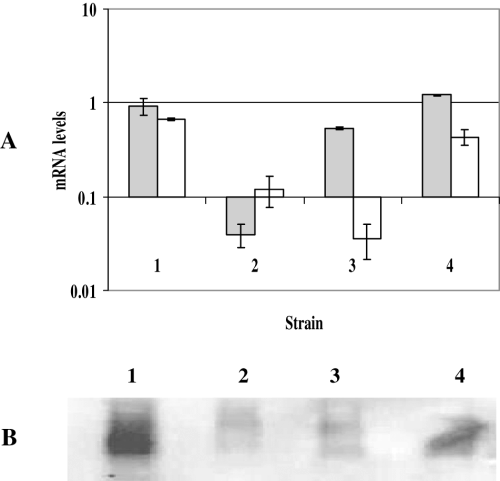
Expression of sigK and Rv0444c in ΔsigK mutant, sigK complement and dyad complement in comparison with the wild-type H37Rv.A. Relative amounts of sigK and Rv0444c mRNA. mRNA levels were measured by SYBR green RT-PCR. Values represent the average of two independent RNA samples and are normalized to sigA mRNA. Grey bars, sigK; white bars, Rv0444c.B. Western blot analysis of the production of Rv0444c-encoded protein in the mutant and complements as compared with H37Rv, using polyclonal rabbit serum raised against recombinant Rv0444c-encoded protein. 1, H37Rv; 2, ΔsigK; 3, ΔsigK::sigK (sigK complement); 4, ΔsigK::dyad (dyad complement).
Expression of mpt70 and mpt83 in ΔsigK and ΔsigK complements as compared with the wild-type strain H37Rv.A. mRNA levels as assayed by RT-PCR coupled with molecular beacon. Grey bars, mpt70; white bars, mpt83.B–D. Western blot analysis of MPT83 (B), MPT70 (C) and MPT64 (D) in ΔsigK and ΔsigK complements. Expression of MPT83 was tested in two clones of ΔsigK::sigK, and ΔsigK::dyad and similar results were obtained for each clone (B). For MPT70 and MPT64 (loading control) expression (C and D respectively), only one clone of ΔsigK::sigK and ΔsigK::dyad was tested. 1, H37Rv; 2, ΔsigK; 3, ΔsigK::sigK; 4, ΔsigK::dyad.
Co-transcription of sigK and Rv0444c and physical interaction of their gene products
For other anti-sigma factors, the genes encoding the anti-sigma factor and the sigma factor are co-transcribed, and there is physical interaction between the anti-sigma factors N-terminus and its cognate sigma factor (Yoshimura et al., 2004; Hahn et al., 2005). To determine whether these properties could be observed for Rv0444c and sigK, first RT-PCR was performed using sigK and Rv0444c primers, providing the expected product (data not shown). Next, analysis of the membrane topology of Rv0444c gene product in silico (http://bioinf.cs.ucl.ac.uk/psipred/) predicted that the Rv0444c-encoded protein would be a transmembrane protein with an intracellular N-terminal and an extracellular C-terminal, consistent with other anti-sigma factors of M. tuberculosis (Hahn et al., 2005). We therefore tested for direct interaction between SigK and the N-terminal part of Rv0444c gene product using the yeast two-hybrid system. As shown in Fig. 7A, the yeast β-galactosidase reporter was activated upon coexpression of SigK and the N-terminal region of Rv0444c, indicating that the proteins can physically interact. Additionally, this transformant grew on media lacking uracil (Fig. 7B), indicating activation of the URA3 reporter gene. Consistent with induction of URA3, the same transformant was unable to grow in the presence of 5-fluoroorotic acid (5FOA), a compound converted to the toxic 5-fluorouracil by uracil (Fig. 7C). In sum, our data indicate that sigK and Rv0444c are co-transcribed, that the N-terminal (cytoplasmic) part of the Rv0444c gene product physically interacts with SigK, and that this interaction results in decreased expression of the SigK-regulated genes (mpt70/mpb83). We therefore refer to the protein encoded by Rv0444c as RskA (regulator of SigK).
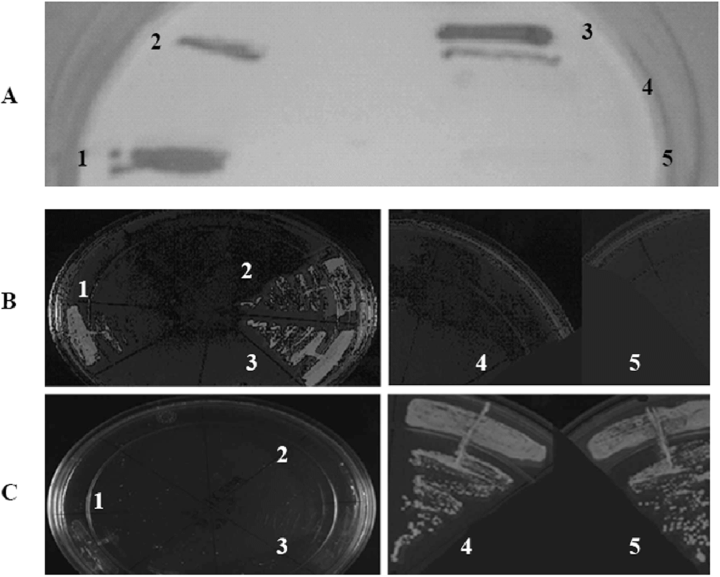
Interaction of SigK and the N-terminal domain of Rv0444c-encoded protein by the yeast two-hybrid system. Interaction is confirmed by increased level of β-galactosidase activity (4A) and growth in the absence of uracil (4B) but not 5FOA (4C). 1, test (transformant with pDEST32 harbouring SigK and pDEST22 harbouring N-terminus of Rv0444c gene product); 2, positive control 1 (control D, ProQuest series 10835); 3, positive control 2 (PQ10001-01); 4, negative control 1 (transformant with bait plasmid harbouring SigK coexpressed with empty prey vector); 5, negative control 2 (transformant with empty bait vector coexpressed with pDEST22 harbouring N-terminus of Rv0444c gene product).
Defining the transcriptional impact of RskA by microarray
To better define the regulon of the SigK-RskA system, we performed two different whole genome microarray analyses. In the first set of microarray experiments, we compared the transcriptional profile of Russia::pMH416 (complement with empty vector) with Russia::Rv0444c (complement with wild-type Rv0444c under the mop promoter). Genes most significantly downregulated upon overexpression of Rv0444c were Rv2873 (mpb83), Rv2875 (mpb70) and Rv2874 (dipZ) (data provided in Table S2). In a second set of microarray experiments, we analysed the transcriptional profile of the ΔsigK mutant against ΔsigK::sigK (single gene complement). In these microarrays, the addition of sigK significantly upregulated expression of Rv0446c and Rv0449c (both part of the sigK locus), and Rv2873 (mpb83), Rv2875 (mpb70) and Rv2876 (Fig. 8). Of note, the only discordance between these two sets of transcriptome analyses was the apparent downregulation of lipP in the sigK complement arrays. As lipP is next to the attB site (where pMV306 integrates), and complementation of BCG Russia employed an extrachromosomal plasmid, we postulate that this result is an artefact of the complementation process, and not that lipP is a SigK-dependent gene. Taken together, these results revealed that the SigK-RskA regulon is small and restricted to two regions: the sigK and the mpt70/mpt83 regions. These results are in agreement with our previous study in which the addition of sigK in a BCG background affected only the expression of the sigK and mpb70/mpb83 regions (Charlet et al., 2005).
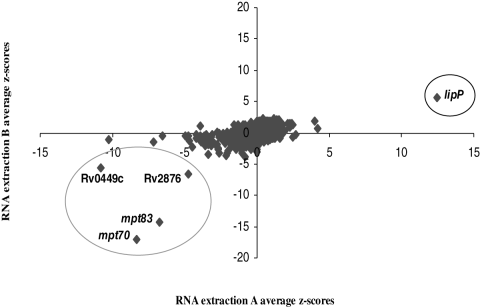
Transcriptional profile of ΔsigK (mutant), and ΔsigK::sigK (sigK complement) by microarray analysis. Genes for which expression was decreased in the ΔsigK mutant as compared with the sigK complement are indicated by a negative Z score and genes with increased expression in the mutant are reported by a positive Z score. Genes whose expression is significantly upregulated by the addition of sigK complement are parts of the sigK and mpt70/83 loci. The only probe presenting downregulation in the sigK complement was for the lipP gene (see text).
Discussion
Although M. tuberculosis and M. bovis share remarkable genomic similarity, phenotypic differences between these two organisms have long been remarked. One of these differences is in the production of the two antigenic proteins MPB70 and MPB83, which are highly expressed in M. bovis and minimally expressed in vitro by M. tuberculosis (Hewinson et al., 1996; Wiker et al., 1996). In this study, we provide evidence that the genetic basis behind this difference is due to variable sequences of a regulatory gene, Rv0444c. We extended this phenotype to other members of the MTC whose levels of MPT70 and MPT83 expression were previously unknown and showed a tight genotype–phenotype correlation between Rv0444c sequence and MPT70/MPT83 expression levels. As in most cases, activity of mycobacterial sigma factors is induced in the presence of a specific signal (Manganelli et al., 2004) and phylognetically, M. tuberculosis presents as ancestral to modern M. bovis (Brosch et al., 2002; Mostowy et al., 2002; 2005), it can be inferred that the inducible phenotype seen in M. tuberculosis is the wild-type scenario whereas the constitutive phenotype in M. bovis is the result of mutations in Rv0444c. It is interesting to note that two different sets of mutations in two different lineages (C320T and C551T in M. bovis and G698C in the Oryx bacillus) led to the same functional consequence, i.e. overproduction of MPT70/MPT83 in vitro. One wonders if these different bacterial lineages have gained a biological advantage by becoming high producers of these two antigenic proteins.
While large genomic deletions initially presented as the most evident form of genetic variability between M. tuberculosis and M. bovis (Behr et al., 1999; Gordon et al., 1999), a number of well-characterized phenotypic differences between these organisms have instead been attributed to SNPs. For instance, unlike M. tuberculosis, M. bovis requires the addition of pyruvate in the culture media for optimal growth, due to a SNP in the pyruvate kinase gene, pykA, that has resulted in an inactive pyruvate kinase in M. bovis (Keating et al., 2005). In another study, the natural resistance of M. bovis to pyrazinamide was shown to be due to a single non-synonymous SNP (C169G) in pncA, resulting in a non-functional pyrazinamidase (Scorpio and Zhang, 1996). By altering the expression profile of two important antigens, this study adds Rv0444c SNPs to the list of M. bovis SNPs that have led to phenotypic differences between this organism and M. tuberculosis.
When compared with the M. tuberculosis lineage, the genome sequence of M. bovis revealed that no genes are unique to M. bovis, leading to the prescient suggestion that differential gene expression due to SNPs in regulatory genes could explain differences in the biology and host preference displayed by these two organisms (Garnier et al., 2003). Additionally, Sreevatsan et al. documented a remarkably restricted polymorphism in structural genes among MTC members and suggested that the rare SNPs observed are generally in regulatory genes and may lead to functional consequences (Sreevatsan et al., 1997). Our laboratory has previously shown that a SNP in the start codon of sigK, a gene encoding one of M. tuberculosis ECF sigma factors, SigK, causes a decrease in the levels of two antigenic proteins MPB70 and MPB83 in BCG strains obtained after 1927 (Charlet et al., 2005). The current study extends this theme by presenting another regulatory gene, Rv0444c, in which SNPs are shown to be functionally significant.
On the basis of these findings and accepted models of Sigma factor regulation, we propose the following model for the regulation of mpt70 and mpt83. In M. tuberculosis H37Rv, the referent wild-type strain, RskA, through its N-terminal domain, binds to SigK and inhibits its activity in vitro. Upon sensing an in vivo stimulus through its extracellular C-terminal domain, RskA releases SigK, freeing it to bind to the promoter of its target genes (mpt70 and mpt83) and initiate their transcription. In M. bovis, RskA is dysfunctional due to at least one of the two SNPs in its encoding gene, Rv0444c, with further site-specific study required to resolve the functional consequence of each mutation. Consequently, in M. bovis, SigK is always ‘on’ and directing transcription of mpt70 and mpt83 in the absence of a stimulus. Anti-sigma factors such as Streptomyces coelicolor RsrA, and M. tuberculosis RshA have been demonstrated to sense and respond to oxidative stresses through regulation of their cognate sigma factors (Kang et al., 1999; Raman et al., 2001). For others, like RslA, the signal and the response are not known but the role of SigL and its regulon in vivo is clearly demonstrated by the significant attenuation of a sigL mutant in mice (Hahn et al., 2005). Similar to RslA, the signal to which RskA senses and responds to is yet to be determined, but it is expected that defining the biological role of the SigK-RskA target genes will provide insight into the in vivo stimuli that activate this system.
Previous data presented by Charlet et al. (2005), and further corroborated in the current study, indicate that SigK-RskA target genes are restricted to just the sigK and mpt70/mpt83 regions. Although it has been reported that ECF sigma factors control a small regulon (Bashyam and Hasnain, 2004), the restriction of the SigK regulon to the mpt70/mpt83 region (besides the sigK locus) is atypical and noteworthy. The regulons of other reported M. tuberculosis ECFs are quite extensive, for instance, microarray analysis indicated that M. tuberculosis SigH positively regulates the expression of 31 genes (Kaushal et al., 2002), SigE controls the expression of about 100 genes, including sigB (Manganelli et al., 2001), and the regulon for SigD includes 47 upregulated genes (Raman et al., 2001). The biological function of MPT70 and MPT83 is still unknown but several lines of evidence strongly suggest their possible role in vivo. First, although in vitro MPT70 and MPT83 protein levels are low in M. tuberculosis, serum from mice infected with live M. tuberculosis react strongly to these antigens (Hewinson et al., 1996). Second, Schnappinger et al. described the transcriptional profile of M. tuberculosis inside stimulated macrophages, and found that mpt70 and mpt83 are both significantly induced in macrophages, with mpt83 also confirmed in infected mice (Schnappinger et al., 2003). Finally, the shut-down in expression of MPB70 and MPB83 by BCG Pasteur argues that there was a cost to producing these proteins in high levels during prolonged in vitro passage in the absence of host pressures. The observation that this regulon has been upregulated in two independent lineages of the MTC further argues towards a yet-to-be determined role of these genes and their protein products in vivo.
By complementing the ΔsigK mutant with sigK alone, we have constructed a M. tuberculosis strain with the M. bovis phenotype for MPB70/MPB83 production. It has been previously reported that M. bovis is more virulent than M. tuberculosis in mice, as demonstrated by increased pathology and disseminated infection induced in the case of M. bovis but not M. tuberculosis (Medina et al., 2006). Additionally, the rabbit model of tuberculosis has historically been used to differentiate between M. bovis and M. tuberculosis, as first described by Theobald Smith, who wrote: ‘The gross results show a sharp demarcation between the bovine and the human cultures. While all the rabbits inoculated with the former [bovine] succumbed in from 17 to 21 days, of the rabbits inoculated with the latter [human] only one succumbed in 35 days’ (Smith, 1898). With this in mind, it shall be interesting to test the mutants generated in these experiments in animal models, to challenge a phenotype first described more than a century ago.
Experimental procedures
Bacterial strains and culture conditions
Mycobacterium africanum 60914 (previously characterized in Mostowy et al., 2004b), the Oryx bacillus 51145, M. caprae 60312 (previously characterized in Mostowy et al., 2005) and M. bovis 62389 are generous gifts from Louise Thibert. M. microti and M. tuberculosis H37Rv are gifts from Dr David Sherman. Mycobacteria and BCG strains were grown at 37°C in Middlebrook 7H9 medium (Difco Laboratories, Detroit, MI) containing 0.05% Tween 80 (Sigma-Aldrich, St Louis, MO) and 10% albumin-dextrose-catalase (Becton Dickinson and Co., Sparks, MD) supplement on a rotating platform (Wheaton). For solid media, Middlebrook 7H10 (without Tween) supplemented with OADC was used. E. coli DH5-α used for cloning purposes was cultured at 37°C in Luria broth (Difco). Antibiotics were added as needed at the following concentrations: kanamycin: 50 μg ml−1 for E. coli and 25 μg ml−1 for mycobacteria; hygromycin: 100 μg ml−1 for E. coli and 50 μg ml−1 for mycobacteria; apramycin: 30 μg ml−1 for both E. coli and mycobacteria.
DNA extraction and sequence analysis
DNA from MTC members was extracted using a protocol based upon lysozyme and proteinase K (Van Soolingen et al., 1991), where samples were used as template for targeted sequence analysis. Primers were designed to amplify Rv0444c and flanking DNA (Rv0444cL: 5′-GGCGCTCATGACTGAACATA-3′ and Rv0444cR: 5′-CTAGTGGTACCCGGCGTGTT-3′) where amplicons were subject to direct dideoxy terminal sequencing at the McGill University and Genome Quebec Innovation Center (http://genomequebec.mcgill.ca/). Sequenced products were analysed by alignment searches against published genome sequences, namely M. tuberculosis H37Rv using Tuberculist (http://genolist.pasteur.fr/TubercuList/), M. tuberculosis 210 and CDC1551 using the sequences provided at NCBI (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi), M. bovis AF2122/97 using Bovilist ( http://genolist.pasteur.fr/BoviList/), and the assembly sequence of BCG Pasteur (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/m_bovis).
Preparation and infection of THP-1 cells
To prepare the human macrophage-derived THP-1 cell line for infection as previously described (Lewis et al., 2003), cells were pelleted by centrifugation, resuspended in RPMI 1640 medium plus 10% fetal calf serum (FCS) with 100 nM phorbol 12-myristate 13-acetate (PMA; Sigma), and delivered (1–10 × 107 cells in 40 ml) into 175 cm2 tissue flasks (Falcon). Stimulated THP-1 cells were additionally provided with human IFN-γ from BioDesign International at this time (100 U ml−1). Cells were incubated at 37°C with 5% CO2 for 24 h. PMA (with or without IFN-γ)-containing medium then was removed from the flasks, cells were washed with warm RPMI 1640 medium, given fresh RPMI 1640 medium plus FCS, and re-incubated for 24 h during the infection process. For each infection, bacteria stocks (M. tuberculosis H37Rv and H37Ra) were grown to mid-log phase in 7H9 media, diluted in warm RPMI 1640 medium plus 10% FCS (THP-1 complete media), and added to each flask containing 1–10 × 107 THP-1 cells at a multiplicity of infection of 5–10. Cells were incubated at 37°C in 5% CO2. After 24 h, extracellular bacteria were removed, and RNA was isolated from internalized bacteria (see below).
Ex vivo RNA isolation
Based on published RNA extraction protocols (Mangan et al., 2002; Schnappinger et al., 2003), RNA was isolated from bacteria within infected macrophages, yielding approximately 1 μg from 107 host cells. Specifically, 24 h after infection, macrophages were lysed in low detergent concentrations in presence of guanidinium isothiocyanate, followed by phagosome isolation and bacterial harvest by ultracentrifugation. Bacterial RNA was subsequently extracted and analysed by qRT-PCR as described below.
Complementation assays
To complement BCG Russia, a 1068 bp DNA fragment containing the Rv0444c gene plus 292 bp upstream was PCR amplified from M. tuberculosis H37Rv or from BCG Russia using the following primers: Rv0444F-XbaI: 5′-ATAAAATCTAGAGGTGCGGCCAACGTCGATC-3′; Rv0444R-HindIII: 5′-ATAAAAGCTTCCGGCGTGTTCGTCGCGATGC-3′. The PCR amplicon was first cloned into pCR 4-TOPO cloning vector (Invitrogen, Carlsbad, CA). The fragment was then removed from the TOPO vector with the restriction enzymes EcoRI (found in the TOPO vector) and HindIII and cloned into pMH416 digested with the same enzymes. pMH416 (a kind gift of D.R. Sherman and M.J. Hickey) is a derivative of pMH29 (George et al., 1995) that has the apramycin-resistance gene (Cangelosi et al., 2006). The resulting plasmid pMH416::Rv0444c as well as the pMH416 empty vector were electroporated into BCG Russia. Complementation was PCR-confirmed and amplicons were sequence-confirmed for all transformants.
Construction of sigK mutant and complemented strains
Mycobacterium tuberculosis H37Rv sigK mutant was created by allelic exchange in a two-step selection process, replacing sigK with a kanamycin cassette. For this purpose, we used the plasmid pKO, which contains the sacB (conferring sucrose sensitivity) and a hygromycin cassette in its backbone as described in Lewis et al. (2003). pKO multiple cloning sites are also flanked by a kanamycin-resistance gene. DNA fragments of 1464 bp and 1666 bp spanning the regions proximal and distal of sigK, respectively, were PCR amplified from H37Rv genomic DNA using the following primers: sigKproxF: 5′-ATAAGCATGCAGCGATGCGTTGGGAGAG-3′; sigKproxR: 5′-ATAAAAGCTTGGATGCAGCTGAGGGTCTG-3′ and sigKdistF: 5′-ATAAGGTACCTAGCCGTGCACTATGACCTG-3′; sigKdistR: 5′-ATAAGGTACCCGATGGGTAGCCTATCGCCA-3′. The PCR amplicons were independently cloned into pCR 4-TOPO cloning vector (Invitrogen, Carlsbad, CA). The distal fragment was digested from TOPO with KpnI and ligated to the pKO vector digested with the same enzyme, making the plasmid pKO-distal. PCR and restrictions digests were used to confirm the presence and the correct orientation of the insert. The proximal region was digested from TOPO with SpHI and HindIII and cloned into pKO-distal plasmid digested with the same enzymes. The final construct pKOSigK contains the regions proximal and distal to sigK on each side of the kanamycin-resistance gene. The construct was confirmed by amplifying and sequencing the two regions (proximal and distal). We then transformed the pKOSigK plasmid into M. tuberculosis H37Rv using the method previously described (Belley et al., 2004). Hygromycin-resistant colonies obtained from the first selection were selected and analysed for site-specific integration using the Expand Long Template PCR System (Roche). The clones in which the plasmids integrated at the right place were plated on 7H10 plus kanamycin and 2% sucrose. Kanamycin- and sucrose-resistant colonies would have lost the plasmid backbone containing sacB (Pelicic et al., 1996). These colonies were tested on hygromycin and kanamycin independently. Those colonies that grew on kanamycin but not on hygromycin were selected for further analysis. sigK deletion was first confirmed by PCR with primers flanking the sigK gene and sequencing, and also confirmed by Southern blot analysis.
To complement the ΔsigK mutant with sigK only, we used the plasmid pH37Rv, which is a derivative of pMV306 containing the sigK region (including the complete gene and 288 bp upstream) as previously described (Charlet et al., 2005). To complement the ΔsigK with the dyad (sigK and Rv0444c), we cloned a 1703 bp PCR fragment containing sigK, Rv0444c plus 288 bp upstream of sigK into pMV306, using the following primers: SigKF-NotI: 5′-ATAAGCGGCCGCACGCGTCACCCCAACTACT-3′; Rv0444R-EcoRV: 5′-ATAAGATATCCCGGCGTGTTCGTCGCGATGC-3′. NotI and EcoRV sites were added to facilitate the cloning of this fragment into pMV306. Because pMV306 originally has a kanamycin-resistance gene and the ΔsigK mutant generated was kanamycin-resistant, we genetically replaced the kanamycin cassette with a hygromycin-resistance marker. The presence of the correct insert was confirmed by restriction digest and PCR followed by sequencing. The resulting plasmids pMV306-hyg::sigK and pMV306-hyg::dyad were electroporated into the ΔsigK mutant. Complementation was PCR-confirmed by amplifying the inserts with primers specific for the regions of pMV306 (Charlet et al., 2005) flanking the insert and amplicons were sequence-confirmed for all transformants.
RNA extraction and RT-PCR
Natural and genetically engineered strains were grown in vitro to an OD600 of 0.2–0.6, where they were pelleted by centrifugation, resuspended in 1 ml of wash buffer (0.5% Tween 80, 0.8% sodium chloride) and transferred to 2.0 ml screw-cap tubes. RNA was isolated by a modified phenol-chloroform extraction protocol as previously described (Charlet et al., 2005). Genomic DNA contamination was removed by RNAeasy on-column digestion, following the manufacturer's protocol (Qiagen, Mississauga, ON). Once the quality of RNA was confirmed by denaturing gel electrophoresis (formaldehyde), it was converted to cDNA using Fermentas cDNA kit.
To determine mRNA levels for mpb70, mpb83 and sigA, we performed qRT-PCR with molecular beacons, applying sigA levels as a normalization control (Manganelli et al., 1999). The qRT-PCR method as well as primers and beacons for sigA, mpb70 and mpb83 used in this study are previously described in Charlet et al. (2005). To determine levels of sigK and Rv0444c as function of our deletion and complementation mutants, we modified this protocol to test expression by qRT-PCR with SYBR green. To test for co-transcription of sigK and Rv0444c, primers within each gene (SigK-CTXL: 5′-GGGCTGACGTATGTCGAAGT-3′ and Rv0444-CTXR: 5′-CTCGTTCATCGTCGGACAC-3′) were employed in PCR analysis of mRNA with and without reverse transcriptase, to demonstrate message bridging the genes.
Protein preparation and immunoblot analysis
For secreted proteins, culture supernatants were filtered with a 0.22 μm membrane filter and concentrated with an Millipore Ultra-10 Centrifugal Filter Unit, 10 000 MW cut-off. For membrane-associated proteins, culture pellet was resuspended in 1 ml PBS and samples were subjected to the Fast Prep (BIO 101 Savant) for 15 s at 6.0 rpm. Samples were incubated on ice for 10 min. Cell debris were removed by spinning down the samples and the supernatant was boiled for 20 min. SDS-PAGE was performed under reducing conditions using the Mini-PROTEAN 3 electrophoresis system (Bio-Rad) with 12% polyacrylamide gels. Proteins were transferred to a polyvinylidene difluoride membrane. Rabbit polyclonal antibodies against MPB70 and MPB83 (gifts of Harald Wiker) were blotted onto the membrane and the ECL Western kit (Millipore, Billerica, MA) was used for protein detection. As internal loading control, the same samples were transferred to a membrane and blotted with a mouse polyclonal antibody against MPT64 (gift of Mark Doherty), a secreted antigen whose expression is not thought to be affected by SigK. To detect levels of Rv0444c-encoded protein, we used rabbit serum raised against recombinant Rv0444c (Chemicon International, Temecula, CA) obtained as follows. Rv0444c was amplified from H37Rv and cloned into the pET21 vector. After sequence confirmation, the plasmid was transformed into E. coli BL21 (DE3) pLysS (Novagen) and the protein expressed through overnight induction with 0.5 mM IPTG. Cells were harvested by centrifugation (5000 g, 10 min and 4°C) and disrupted by sonication at 4°C. Cell debris was removed by centrifugation, and the supernatant was loaded onto a 1 ml HiTrap chelating column (Amersham Biosciences). The column was extensively washed with loading buffer and eluted using a 0–600 mM imidazole gradient in loading buffer to reveal a single band of the expected length on SDS-PAGE gel. This product was then sent for preparation of serum.
Yeast two-hybrid analysis
Protein interaction was assayed using the ProQuestTM two-hybrid system (Invitrogen, Carlsbad, CA). By recombination cloning, we engineered a bait plasmid designed to overexpress recombinant SigK linked at its N-terminus to the Gal4 DNA-binding domain (pDEST32). Similarly, the prey plasmid expressed the N-terminal part of Rv0444c (amino acid residues 1–92) fused to GAL4 activation domain (pDEST22). pDEST32 and pDEST22 contain TRP1 and LEU2, respectively, permitting selection on media lacking tryptophan or leucine (synthetic complete media leu–trp–). Bait and prey plasmids were transformed into the mating-type yeast strain MAV203 using the method essentially described by Gietz et al. (1992). Briefly, yeast strains were grown in YPAD medium to an OD600 of ∼1. Competent cells were prepared with 1× TE/LiAc solution and transformation was carried out by mixing yeast cell suspension with 50 μg of single stranded salmon sperm DNA, plasmid DNA (and 0.3–1 μg) and 40% PEG. Cells were incubated at 30°C with agitation (roller drum) for 30 min, followed by a heat shock at 42°C for 15 min. The MAV203 yeast strain contains integrated reporter genes such as ura3 and lacZ and positive interactions are detected when the GAL4 binding and activation domains are bridged via a direct protein–protein interaction, and activation of the reporter genes. Transformants were plated on SC-L-T-Ura and 5FOA plates to test for growth. For the β-galactosidase assay, transformants were grown overnight on nitrocellulose filter paper placed on YPAD agar plate as described in the invitrogen ProQuest Two-hybrid system manual. The filter was soaked in Z-buffer containing β-ME and Xgal after rapid exposure to liquid nitrogen and incubated overnight on Whatman paper.
Microarray analysis
Microarray hybridization and analysis were performed as previously described (Charlet et al., 2005). In brief, mRNA from BCG strains and complemented strains was extracted during log-phase in vitro growth and labelled with Cy3 or Cy5 dUTP by reverse transcriptase (Amersham Biosciences). Labelled cDNA was hybridized to microarrays composed of oligonucleotide probes from the TB Array-Ready Oligo SetTM (Operon) printed onto SigmascreenTM microarray slides (Sigma). Initial comparisons were M. tuberculosis H37Rv versus select MTC members (Table S1). After complementing BCG Russia with wild-type Rv0444c, comparisons of Russia::pMV306 versus Russia::Rv0444c were also performed (Table S2). Hybridized arrays were scanned with ScanArray 5000XL and hybridization results were quantified with ScanArray software (PerkinElmer, Freemont, CA). Array analysis was performed as previously described (Mostowy et al., 2004c; Charlet et al., 2005) and can be summarized as such. Subtracting total spot intensity minus the surrounding background produced a corrected spot intensity. Negative corrected spot intensities were set to +1. All spots flagged as misrepresentative (array artefacts) or unrecognizable (quiescent in both channels) by ScanArray were analytically ignored. Intensity ratios (Cy3/Cy5 or Cy5/Cy3) were determined using corrected spot intensities and log10 transformed. Values for each gene were obtained in duplicate for each array (inherent to array design) and averaged. For each array, a representative Z-score, indicative of how many standard deviations a data point lies from the population mean, was calculated for each gene. Z-scores for each gene were determined across replicates within each experiment to minimize the probability of observing such variation by chance alone. Genes with Z-scores of two or greater across arrays are presented.
Acknowledgements
The authors thank the following people for their input and help with experiments: Danielle Charlet, Elizabeth Fidalgo, Frédéric Veyrier, Pierre Colas, Sarah Sanowar, David Alexander, Fiona McIntosh, Robert Kozak, Francois Coulombe, David Sherman and members of the Schurr laboratory. BSS received a fellowship from the MUHC Research Institute. S.M. was funded by a studentship of the Fonds de la Recherche en Santé du Québec (FRSQ). M.B. is Chercheur Boursier Senior of the FRSQ. Work was funded by an operating grant from the Canadian Institutes for Health Research, MOP-79309.