Widespread distribution of a lexA-regulated DNA damage-inducible multiple gene cassette in the Proteobacteria phylum
Summary
The SOS response comprises a set of cellular functions aimed at preserving bacterial cell viability in front of DNA injuries. The SOS network, negatively regulated by the LexA protein, is found in many bacterial species that have not suffered major reductions in their gene contents, but presents distinctly divergent LexA-binding sites across the Bacteria domain. In this article, we report the identification and characterization of an imported multiple gene cassette in the Gamma Proteobacterium Pseudomonas putida that encodes a LexA protein, an inhibitor of cell division (SulA), an error-prone polymerase (DinP) and the alpha subunit of DNA polymerase III (DnaE). We also demonstrate that these genes constitute a DNA damage-inducible operon that is regulated by its own encoded LexA protein, and we establish that the latter is a direct derivative of the Gram-positive LexA protein. In addition, in silico analyses reveal that this multiple gene cassette is also present in many Proteobacteria families, and that both its gene content and LexA-binding sequence have evolved over time, ultimately giving rise to the lexA lineage of extant Gamma Proteobacteria.
Introduction
Bacterial cells contain several pathways involved in DNA repair. Although most of them target specific DNA lesions (i.e. oxidative damage or presence of alkyl radicals in DNA), a global DNA damage response system is present in most bacteria. The LexA-mediated SOS response, first described and thoroughly studied in Escherichia coli (Walker, 1984), is addressed to guarantee cell survival when massive DNA damage is introduced and the normal DNA replication of the bacterial cell is disturbed. The E. coli SOS network comprises at least 40 genes that are under direct control of the lexA and recA genes, which are also members of this network (Fernández de Henestrosa et al., 2000; Courcelle et al., 2001; Khil and Camerini-Otero, 2002). Many of the E. coli SOS genes have been associated to a particular cellular process, such as transitory inhibition of cell division (sulA), error-prone replication (umuDC and dinP) or excision repair (uvrAB). In the absence of DNA injuries, the E. coli LexA protein binds specifically to a regulatory motif with consensus sequence CTGTN8ACAG (the E. coli SOS box), thereby effectively blocking transcription of SOS genes (Walker, 1984). Conversely, in the advent of DNA damage the RecA protein acquires an active conformation (RecA*) after binding to single-stranded DNA fragments generated either by DNA damage-mediated replication inhibition or by enzymatic processing of broken DNA ends (Sassanfar and Roberts, 1990). Upon activation, RecA* promotes autocatalytic cleavage of the Ala84-Gly85 bond of E. coli LexA (Little, 1991). This cleavage, mediated by LexA residues Ser119 and Lys156, is similar in mechanism to that observed for serine proteases (Little, 1991; Luo et al., 2001) and effectively prevents LexA from binding SOS regulatory motifs, thus resulting in a global induction of the SOS response. After removal of DNA lesions, RecA* concentration declines and non-cleaved LexA protein returns to its usual levels, inhibiting again the expression of SOS genes.
The LexA protein is, with some notable exceptions, widely distributed across the Bacterial Domain. Presence of the E. coli SOS box in the promoter region of DNA damage-inducible genes has been described in several families of the Gamma Proteobacteria, such as Pseudomonaceae, Aeromonadaceae, Vibrionaceae and Pasteurelaceae, and even in some Beta Proteobacteria (e.g. Ralstonia solanacearum, Bordetella bronchiseptica, Bordetella parapertussis or Burkholderia cepacia). Moreover, in silico analyses have shown that LexA controls a regulon of 10–40 genes in these species (Erill et al., 2003). However, the E. coli LexA-binding sequence is not preserved in other bacterial groups where LexA-dependent DNA damage-inducible activity has been reported. For instance, Alpha Proteobacteria possess a markedly divergent LexA-binding motif, with a GTTCN7GTTC direct repeat consensus sequence that is monophyletic for this class (Fernández de Henestrosa et al., 1998; Tapias and Barbé, 1999). Similarly, all Gram-positive bacteria studied so far display a highly conserved LexA recognition motif with the CGAACRNRYGTTYC consensus sequence (Winterling et al., 1998; Davis et al., 2002). Nevertheless, this motif is not exclusive of Gram-positive bacteria, because it has been demonstrated to be also the LexA-binding motif in some Gram-negative phyla, such as the Green non-sulphur bacteria (Fernández de Henestrosa et al., 2002). Likewise, the Cyanobacteria LexA-binding sequence (RGTACNNNDGTWCB) seems to be a direct derivative of the Gram-positive one (Mazón et al., 2004).
Even though it is not the usual situation, duplicity of SOS regulatory genes has been described for a number of bacteria spanning different phyla. Myxococcus xanthus, for instance, presents two copies of the recA gene, but only one of these two recA genes (recA1) is DNA damage inducible (Norioka et al., 1995). Even so, in the promoter region of the M. xanthus recA2 gene, a mutant derivative sequence of the M. xanthus LexA-binding motif is still present (Campoy et al., 2003). In a similar case, Geobacter sulphurreducens presents two copies of the lexA gene, encoding each a functional LexA protein able to recognize the same DNA binding sequence in both promoters: GGTTN2CN4GN3ACC (Jara et al., 2003). Although both these instances of gene duplicity could be attributed either to a gene duplication event or to lateral gene transfer (LGT), the close similarities between the duplicated proteins and their LexA-binding sites suggest that this duplicity is a consequence of the former.
In contrast, the Gamma Proteobacteria Pseudomonas putida presents a case of lexA duplicity that cannot be readily explained as a consequence of a gene duplication event. It has been demonstrated that P. putida has a lexA gene (lexA1) whose product binds an E. coli-like SOS box (Calero et al., 1991) and that is placed upstream of a sulA-like (sulA1) gene (Weinel et al., 2002). Recently, however, complete sequencing of the P. putida genome has revealed the presence of a second lexA gene (lexA2) in this organism (Weinel et al., 2002), but in silico screening of the whole P. putida chromosome has shown that this lexA2 gene does not present any E. coli-like SOS motifs in its promoter sequence (Erill et al., 2003). In the present work, this lexA2 gene of P. putida has been cloned, and its encoded product has been characterized to analyse both its origin and its relationship with the LexA protein of other bacterial species. The results here obtained demonstrate that the P. putida lexA2 gene is the first gene of an imported polycistronic transcriptional operon to which three other genes involved in DNA repair belong and whose expression is under direct control of its own product: the LexA2 protein. Furthermore, this operon and closely related derivatives are widespread in the Proteobacteria Phylum.
Results
Transcriptional organization of the P. putida chromosomal region containing the lexA2 gene
Immediately downstream the lexA2 gene (PP3116), there are three ORFs (PP3117, PP3118 and PP3119) that have been annotated as sulA, dinP and dnaE genes (Fig. 1A) in the published complete genome of P. putida (Weinel et al., 2002). As it is well known, these genes encode, respectively, a cell division inhibitor, the DNA polymerase IV and the alpha subunit of the DNA polymerase III. To determine whether the lexA2 gene is co-transcribed with these ORFs, a reverse transcriptase polymerase chain reaction (RT-PCR) analysis was performed using RNA extracted from P. putida cells. Results indicate that a lexA2-sulA2-dinP-dnaE transcript is definitely present in P. putida (Fig. 1B), showing that these four genes constitute a single polycistronic unit. Furthermore, it was assessed that expression of this transcriptional unit increases when cells are treated with DNA damaging agents such as mitomycin C (Fig. 2). However, RT-PCR experiments using the oligonucleotides shown in Fig. 1A demonstrate that, in a lexA2(Def) mutant, lexA2 promoter transcription does not increase due to DNA damage (Fig. 2). Moreover, in lexA2(Def) cells the lexA2 promoter presents a basal expression that is practically equal to that obtained in mitomycin C-treated wild-type cells (Fig. 2). Besides, and as expected because of the insertion of the ΩKm interposon inside the lexA2 gene coding region, a lexA2-sulA2-dinP-dnaE transcript cannot be detected in the lexA2(Def) mutant (data not shown). Taken together, these results demonstrate that the lexA2 gene product regulates the DNA damage-mediated expression of the whole lexA2-sulA2-dinP-dnaE operon, and that no additional internal promoters are present in this transcriptional unit.
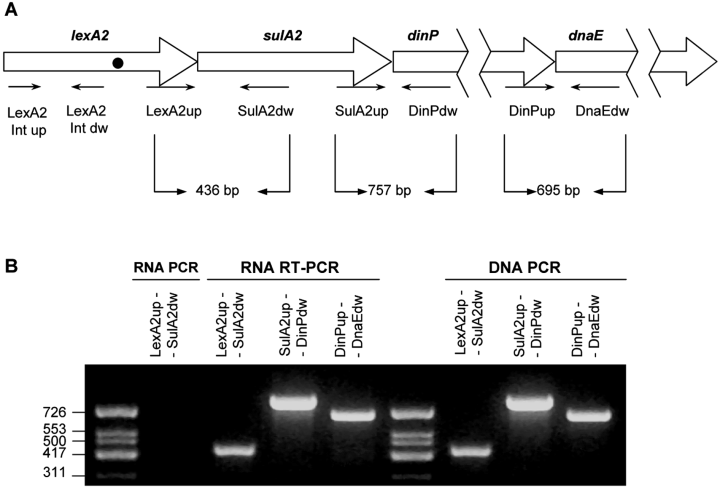
A. Structural arrangement of P. putida lexA2-sulA2-dinP-dnaE genes. Arrows denote the primers used to determine transcripts. The black dot in the lexA2 gene indicates the insertion point of the Ω-interposon (Int). Arrows upstream this point mark the position of primers used to measure the lexA2 gene expression. B. RT-PCR analysis of lexA2-sulA2-dinP-dnaE transcripts present in total RNA from P. putida (RNA RT-PCR). As a control, PCR experiments were also carried out using either DNA (DNA-PCR) or RNA (RNA-PCR) as a template. The molecular mass marker (HinfI-digested DNA of φx174) size is shown at left in base pairs.
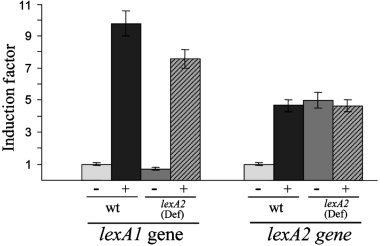
Mitomycin C-mediated induction of P. putida lexA1 and lexA2 genes measured by quantitative RT-PCR in wild-type (wt) and lexA2(Def) cells. The induction factor is the ratio of the relative mRNA concentration for each gene to that of mitomycin C-untreated wild-type cells. The relative mRNA concentration for a given gene is its amount normalized to that of the P. putida trpA gene. Values were calculated 2 h after mitomycin C addition. In each case, the mean value from three independent experiments (each in triplicate) is shown. Symbols (+) and (–) represent addition or not of mitomycin C (20 µg ml−1) respectively.
Identification of the P. putida LexA2-binding sequence
Data presented in Fig. 2 indicate that the LexA2 protein negatively regulates the P. putida lexA2-sulA2-dinP-dnaE operon. To identify the LexA2 recognition sequence, electrophoresis mobility shift assays (EMSAs) were carried out with this purified protein and using as a probe a DNA fragment extending from −140 to +72 of the P. putida lexA2 gene promoter (with respect to its putative translational starting point). The addition of LexA2 protein, but not that of LexA1, specifically decreased the mobility of the lexA2 promoter, as an excess of unlabelled lexA2 promoter abolished it. However, the same did not occur when an excess of non-specific DNA was added (Fig. 3A). These data, together with those concerning the gene expression profiles presented in Fig. 2, clearly demonstrate that lexA1 and lexA2 genes do not present cross-regulation.
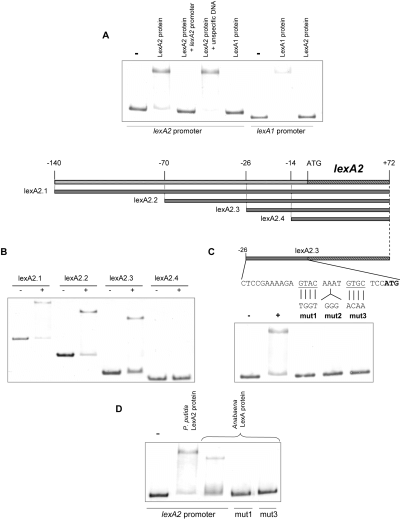
A. EMSAs of the P. putida lexA1 and lexA2 promoters in presence of 80 ng of purified LexA1 or LexA2 proteins. When necessary, a 300-fold molar excess of either unlabelled lexA2 promoter or pGEM-T plasmid DNA was used as a, respectively, specific or unspecific competitor fragment. B. Electrophoretic mobilities of several lexA-promoter fragments in presence (+) or absence (–) of the LexA2 protein (80 ng). C. Effect of the substitution of either GTAC or GTGC motifs, as well as of the insertion of three nucleotides (GGG) between them, in the electrophoretic mobility of the LexA2.3 fragment in presence of LexA2 protein. D. Binding of the purified Anabaena sp. LexA protein to the wild-type or mutant derivatives of the P. putida LexA2.3 fragment. As a control, the binding of the P. putida LexA2 protein to the wild-type DNA fragment is shown.
To define precisely the P. putida LexA2 binding site, serial deletions of the upstream promoter region of the lexA2 gene were generated and analysed in EMSAs with the purified LexA2 protein. Results indicate that the LexA2-binding motif is located between the −26 and −14 positions upstream lexA2 translational start codon (Fig. 3B). A further analysis of this region revealed the presence of the AGTACAAATGTGCTCC sequence, which is very similar to the recently reported LexA-binding motif RGTACNNNDGTWCB of the Cyanobacteria Phylum (Mazón et al., 2004). Several mutations were introduced in the AGTACAAATGTGCTCC sequence to confirm that this was the P. putida LexA2 box. As it can be seen in Fig. 3C, these mutations inhibited the binding of the LexA2 protein to the lexA2 promoter. In accordance with these facts, the Anabaena LexA protein is able to bind to the P. putida wild-type lexA2 promoter but not to its derivative mutants (Fig. 3D), implying that the Cyanobacterial and the P. putida LexA2 binding sites are closely related.
Distribution of the lexA-sulA-dinP-dnaE transcriptional unit in the Bacteria Domain
The results shown above suggested a close relationship between the P. putida lexA2 and the cyanobacterial lexA gene products. Considering the evolutionary gap between both phyla, this resemblance prompted the possibility that the lexA2-sulA2-dinP-dnaE transcriptional unit had been acquired by P. putida through LGT. To explore this possibility, a tblastn search for each one of the four proteins encoded by the lexA2-sulA2-dinP-dnaE operon was carried out against all the available sequenced bacterial genomes. The lexA2-sulA2-dinP-dnaE operon was not found in any of the Gram-positive bacteria and cyanobacteria genomes analysed. However, either this transcriptional unit or others closely related to it were detected in several Gram-negative bacteria belonging to different Proteobacteria classes (Fig. 4). Two major differences could be identified among the detected cassette orthologues: (i) the nature of their regulatory sequences and (ii) the loss of some of the genes belonging to them. Thus, three different instances of the original cassette, besides isolated lexA, sulA, dinP and dnaE genes, were identified: the own lexA-sulA-dinP-dnaE structure, and sulA-dinP-dnaE and lexA-sulA cassettes.
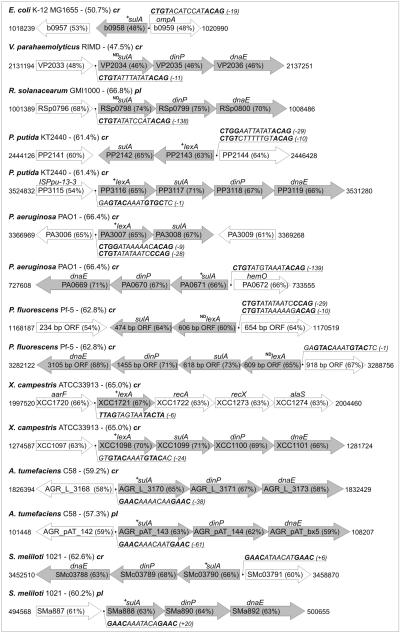
Genetic arrangement of representative lexA-sulA-dinP-dnaE cassettes and their derivatives. Putative and reported LexA-binding sites are displayed with their distance to the translational start codon. The + sign preceding the operon heading gene indicates experimentally confirmed binding of the corresponding LexA protein and DNA damage induction of the operon either in this work (A. tumefaciens, P. putida, P. aeruginosa, S. meliloti and X. campestris) or previously (E. coli). In turn, ND designates non-determined induction/binding ability. Gene G+C percentages for comparison with the overall G+C content of each bacterium, as well as accession numbers, are shown. cr and pl are contractions of, respectively, chromosome and plasmid molecules. Shading denotes cassette-related genes. P. fluorescens positions are relative to the origin of TIGR_220664|contig:3337 fragment of its unfinished genome (NC_004129).
The distinct dissemination of the identified cassette across several Proteobacteria classes is indicative of a complex evolutionary history. On the one hand, a sulA-dinP-dnaE cassette is present in all the Alpha Proteobacteria genomes whose complete sequence has been published to date. Particularly, in Agrobacterium tumefaciens and Sinorhizobium meliloti, where the sulA-dinP-dnaE cassette is present both in plasmids and in the chromosome, the detected cassettes also form a single transcriptional unit that has been shown to be damage inducible (Fig. 4). Moreover, a classical Alpha Proteobacteria LexA SOS box (GTTCN7GTTC) is present in the promoter region of these cassettes, to which the S. meliloti LexA protein binds specifically (Fig. 4). On the other hand, and in contrast with the monophyletic nature of the sulA-dinP-dnaE cassette in the Alpha Proteobacteria, the Gamma Proteobacteria class presents a noticeably larger heterogeneity. Aside from P. putida, the lexA-sulA-dinP-dnaE operon is present in other phytopathogenic pseudomonads (e.g. Pseudomonas fluorescens and Pseudomonas syringae) as well as in Xanthomonas campestris and Xanthomonas axonopodis. It must be noted that, in all these instances, the DNA binding sequence of the P. putida LexA2 protein (AGTACAAATGT GCTCC) is present in the promoter region of the cassette lexA gene. In fact, the purified product of the lexA gene of the X. campestris lexA-sulA-dinP-dnaE operon is able to bind both its own promoter and the P. putida lexA2 one (data not shown). Furthermore, and as in the case of P. putida, all these Pseudomonas species contain also an additional lexA-sulA operon that presents the E. coli-like LexA-binding sequence (CTGTN8ACAG) in its promoter. In contrast, the lexA gene of X. campestris and X. axonopodis presents its own regulatory motif (TTAGN6TACTA) and is not placed upstream the sulA gene, but immediately before recA (Campoy et al., 2002; Yang et al., 2002).
Still, other Gamma Proteobacteria species display additional cassette organizations. The Alteromonadaceae Shewanella oneidensis and the animal-pathogenic Pseudomonas aeruginosa, for instance, present only one lexA gene with the same lexA-sulA arrangement of P. putida lexA1 (Fig. 4). However, P. aeruginosa also possesses a DNA damage-inducible sulA-dinP-dnaE cassette with an E. coli-like LexA-binding sequence to which P. putida LexA1 is able to bind (Fig. 4). An equivalent sulA-dinP-dnaE gene cassette, with its corresponding E. coli-like LexA-binding motif, is also found in the Beta Proteobacteria R. solanacearum, B. bronchiseptica and B. parapertussis, and in some Vibrionaceae, such as Vibrio parahaemolyticus and Vibrio vulnificus (Fig. 4). Nevertheless, lexA does not conform a tandem with a sulA gene in these five species. It must be noted that, as in A. tumefaciens and S. meliloti, the cassette of R. solanacearum is also located in a plasmid although, in this case, there is not an additional copy in the chromosome (Fig. 4).
Phylogenetic analysis of the lexA-sulA-dinP-dnaE cassette
To shed more light on the basis of this variability in gene content of the lexA-sulA-dinP-dnaE operon, separate phylogenetic analyses were performed for each of the proteins encoded by the genes that belong to the original cassette. As seen in Fig. 5A, Gram-positive, Cyanobacteria and Alpha Proteobacteria LexA proteins group in three independent clusters, although there is a significant relationship between them. However, the rest of Proteobacteria LexA proteins analysed, regardless of their genetic organization in either lexA-sulA-dinP-dnaE operons or as single genes in the bacterial chromosome, clustered all in a separate group. Furthermore, a detailed analysis of this cluster clearly indicates that isolated lexA genes, as well as those belonging to the lexA-sulA-dinP-dnaE cassette, diverged all from a common precursor that evolved separately from the lexA gene of a Gram-positive descendant.
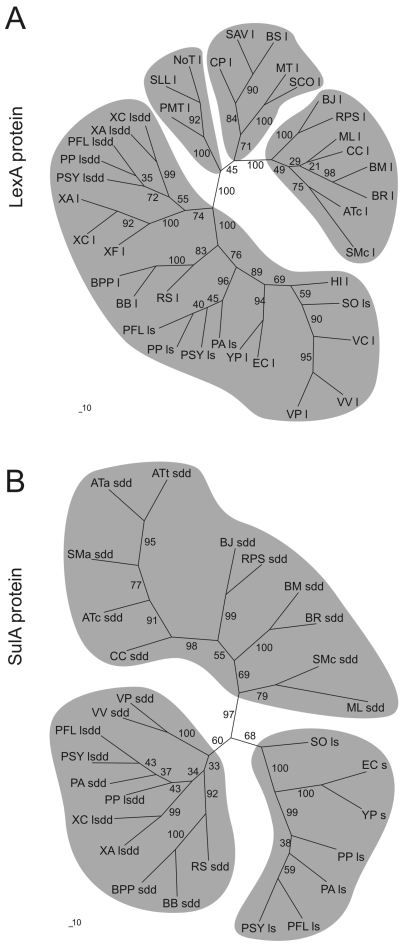
LexA (A) and SulA (B) protein sequence phylogenetic trees. Single l and s characters following the bacterium name represent isolated lexA and sulA genes. Likewise, ls, sdd and lsdd suffixes stand for, respectively, lexA-sulA, sulA-dinP-dnaE and lexA-sulA-dinP-dnaE cassette arrangements. AT, A. tumefaciens; BS, Bacillus subtilis; BB, B. bronchiseptica; BPP, B. parapertussis; BJ, Bradyrhizobium japonicum; BM, Brucella melitensis; BR, Brucella suis; CC, Caulobacter crescentus; CP, Clostridium perfringens; EC, E. coli; HI, H. influenzae; ML, Mesorhizobium loti; MT, M. tuberculosis; NoT, Nostoc; PMT, Prochlorococcus marinus; PA, P. aeruginosa; PFL, P. fluorescens; PP, P. putida; PSY, P. syringae; RS, R. solanacearum; RPS, Rhodopseudomonas palustris; SCO, S. coelicolor; SO, S. oneidensis; SM, S. meliloti; SAV, Staphylococcus aureus; SLL, Synechocystis; VC, Vibrio cholerae; VP, V. parahaemolyticus; VV, V. vulnificus; XA, X. axonopodis; XC, X. campestris; XF, X. fastidiosa; YP, Y. pestis. Characters adjacent to the bacterium name indicate chromosome c or plasmids (a or t ; pSymA, AT or Ti) in those species with cassettes present in both molecules. Numbers at branch nodes indicate bootstrapping values for 100 bootstrap replicates.
Regarding SulA proteins (Fig. 5B), those that are not contained in lexA-sulA-dinP-dnaE or sulA-dinP-dnaE cassettes clustered all in a same group, regardless of their genetic organization either in lexA-sulA tandems or as independent sulA genes. Moreover, Gamma Proteobacteria SulA proteins encoded by either lexA-sulA-dinP-dnaE or sulA-dinP-dnaE operons formed a separate group that included also the Beta Proteobacteria SulA. Less related to both these groups, the Alpha Proteobacteria SulA proteins, which are encoded by a sulA-dinP-dnaE type cartridge, constituted a third cluster, suggesting that this cassette was acquired independently of the one incorporated by Gamma Proteobacteria. The analysis of both DinP and DnaE proteins (data not shown) yielded very similar results, thus validating the above-described scenarios. It is worth noting, however, that Mycobacterium tuberculosis andStreptomyces coelicolor both possess dinP (Brooks et al., 2001; Bentley et al., 2002) and DNA damage-inducible dnaE genes (Flett et al., 1999; Boshoff et al., 2003) whose products are much more closely related to those of the lexA-sulA-dinP-dnaE and sulA-dinP-dnaE cassettes than to their own housekeeping copies of dinP and dnaE. This suggests that both these Gram-positive species could have acquired these dinP and dnaE genes through LGT.
Even so, the analysis of the G+C content of lexA-sulA-dinP-dnaE and sulA-dinP-dnaE operons indicates that, in all the species here analysed, these multiple gene cassettes must have been incorporated early in the speciation process of each organism. This is deduced from the fact that there is a large variation in the mean G+C percentage among detected cassettes (from 45 to 75%) while, for each case, the cassette G+C content is very close to that of the species containing it (Fig. 4).
Discussion
Evidence for lateral gene transfer of a lexA gene in the Proteobacteria
In the present work we have demonstrated that P. putida contains two functional and independent lexA genes. One of them (lexA1) constitutes a lexA1-sulA1 tandem operon harbouring an E. coli-like LexA-binding sequence in its promoter. The second lexA (lexA2) belongs to a multiple gene cassette with the following gene order: lexA2-sulA2-dinP-dnaE. This cassette has been shown to be a DNA damage-inducible transcriptional unit and, in fact, the LexA2 protein controls the expression of its encoding operon by binding to the AGTACAAATGTGCTCC recognition sequence present in its promoter (Fig. 3C). Interestingly, this motif is also able to bind the Cyanobacteria LexA protein (Fig. 3D). Furthermore, we have also shown that the lexA-sulA-dinP-dnaE gene cassette and its derivatives are widespread among the Proteobacteria Phylum (Fig. 4). Taken together, these facts convey strong evidence that LGT events involving the lexA gene have taken place at least once in the evolutionary history of the Gamma Proteobacteria.
With the available data, the exact moment and processes that gave rise to the original lexA-sulA-dinP-dnaE gene cassette here reported are difficult to pinpoint. Nonetheless, the data here obtained provide substantial insights on the probable origins of this cassette. Given the phylogenetic distance between both phyla, the ability of cyanobacterial LexA protein to bind the P. putida cassette binding sequence suggests at first instance that P. putida acquired the cassette through LGT from the Cyanobacteria. However, none of the available cyanobacteria genomes presents a lexA-sulA-dinP-dnaE gene cassette. Therefore, the donor of this operon to P. putida may have been a cyanobacterium whose genome has not been sequenced yet or another bacterial species that, in agreement with the recently proposed branching order of the bacterial phylogenetic tree (Gupta and Griffiths, 2002), appeared closely after the Cyanobacteria. Thus, it seems probable that this multiple gene cassette was formed de novo in an unknown Gram-negative ancestor of the Gamma Proteobacteria class through genetic reorganization involving extant and/or recently imported genes. In this respect, it should be noted that this precursor presented a LexA protein with a binding sequence closely related to that of both Gram-positive and Cyanobacteria, thereby implying direct inheritance of the LexA protein from Gram-positive bacteria or an immediate descendant. Similarly, dinP and dnaE homologues are present in Gram-positive bacteria, Cyanobacteria and Alpha Proteobacteria, making it very probably that these genes were also vertically transmitted to this unknown ancestor of Gamma Proteobacteria. A sulA gene, however, has not been reported to date in any of these three lineages. Thus, the cassette sulA gene must have been acquired by this Gamma Proteobacteria ancestor from an unknown source. Although LGT from a non-described bacterium is possible, the most plausible origin of the sulA gene lies in either bacteriophages or plasmids. Even if sulA genes have not been previously described in plasmids or phages, it is well known that these extrachromosomal elements frequently encode proteins that interfere with such basic cellular processes as division, gene transcription or DNA replication (Miki et al., 1984; Conter et al., 1996; Goyal et al., 1996; Couturier et al., 1998). Additional support for this hypothesis stems from experimental knowledge of proteins with a similar function in extant phages. The protein encoded by the kil gene of the E. coli defective prophage rac, for instance, inhibits the same target as the SulA protein: the FtsZ protein of E. coli, involved in septum formation (Bi and Lutkenhaus, 1993; Conter et al., 1996).
Relationship between the lexA-sulA-dinP-dnaE cassette and the Proteobacteria lexA genes
LexA protein phylogenetic data (Fig. 5A) indicate that, after its constitution, the lexA-sulA-dinP-dnaE cassette evolved through vertical transmission to give rise to the lexA lineage of extant Gamma Proteobacteria. From this point onwards, a deletion of the cassette dinP and dnaE genes probably originated the lexA-sulA tandem that is still conserved in the Pseudomonadacea family and in S. oneidensis. In turn, later deletions or genetic reorganizations of the sulA gene would have given rise, respectively, to the ultimate loss of this gene (such as in the chromosome of Haemophilus influenzae or R. solanacearum) or to the dissociation of the lexA-sulA tandem seen today in E. coli and close relatives (Yersinia pestis, Salmonella typhi, Salmonella typhimurium andShigella flexneri ).
In contrast to the vertical transmission scenario outlined above, the presence in the phytopathogenic pseudomonads and in X. campestris and X. axonopodis of a full lexA-sulA-dinP-dnaE cassette that is markedly divergent from the E. coli-like lexA-sulA tandem also present in the pseudomonads implies that either a duplication or a LGT event took place in an ancestor of these species. At first instance, the high adaptation of the cassettes G+C content seems to support a gene duplication hypothesis, but the divergent LexA-binding motifs of vertically transmitted and cassette lexA genes suggest otherwise (Fig. 4). In addition, the fact that Cyanobacteria LexA protein is still able to bind the LexA-binding motif of both P. putida and X. campestris cassettes (3, 4), the evidence that all lexA-sulA-dinP-dnaE cassettes present self-regulation and the scattered distribution of the cassettes among Gamma Proteobacteria (Fig. 4) all pile up in support of an LGT scheme. Furthermore, when considering an early acquisition scenario, the accepted time since speciation of bacteria with and without cassettes is ample enough for complete amelioration of the cassette G+C content (Koski et al., 2001). Therefore, it seems evident that some time after the lineage presenting the E. coli-like LexA-binding sequence was unambiguously formed, one of its early members acquired by LGT a novel, non-evolved copy of the lexA-sulA-dinP-dnaE cassette (i.e. the lexA2-sulA2-dinP-dnaE cassette here reported for P. putida).
In turn, the presence of a sulA-dinP-dnaE cassette in the closely related P. aeruginosa suggests that the further evolution of this lexA2-sulA2-dinP-dnaE cassette was marked by a deletion of its lexA gene and the emergence of an E. coli-like LexA-binding motif in the promoter region of the sulA2 gene. Thereafter, the presence of several copies of this same sulA-dinP-dnaE operon under E. coli-like LexA control in other Gamma Proteobacteria (V. parahaemolyticus, V. vulnificus) and some Beta Proteobacteria (R. solanacearum, B. parapertussis, B. bronchiseptica) can be explained by reiterated and independent lateral transfer from P. aeruginosa. Likewise, the existence of a sulA-dinP-dnaE operon in the Alpha Proteobacteria could be explained by a similar lateral transfer process: reception of the lexA-sulA-dinP-dnaE cassette, subsequent loss of the governing lexA gene, generation of the necessary Alpha LexA binding box upstream of the sulA-dinP-dnaE operon and plasmid-mediated dissemination. In this respect, it must be noted that similar cases of reiterated lateral transfer have been proposed to explain, for instance, the evolution and organization of the tryptophan biosynthetic genes in the Bacteria Domain (Xie et al., 2003). The fact that sulA-dinP-dnaE cassettes regulated either by the Alpha or by the E. coli-like LexA repressor (Fig. 4) can be found in plasmids both in the Alpha Proteobacteria and in the Beta Proteobacterium R. solanacearum gives further support to the hypothesis of LGT in both classes.
Finally, LexA phylogenetic evidence in the xanthomonads (Fig. 5A) suggests that their conserved lexA-recA tandem is not a product of the evolution of the original lexA-sulA-dinP-dnaE cassette but a duplication of the later acquired, non-evolved cassette that, in the case of Xylella fastidiosa, seems to have been lost as a consequence of a phage-mediated rearrangement still evident in its genome sequence (Simpson et al., 2000). The fact that the surrounding makeup of the Xanthomonas and Xylella recA gene (Fig. 4) preserves the typical Gamma Proteobacteria structure, recX and alaS genes downstream recA (De Mot et al., 1994), further supports the hypothesis of a gene duplication event with insertion of the lexA gene upstream the recA-recX transcriptional unit.
Thus, the results here reported convey a consistent outline of the evolutionary process of the described lexA-sulA-dinP-dnaE cassette and of its influence on the evolution of the SOS system of Gamma Proteobacteria. It seems evident, though, that the availability of more sequence data across different bacterial phyla will make possible to pin down the exact branching point in evolution and, possibly, the precise species in which the lexA-sulA-dinP-dnaE operon here identified was formed, and will also shed light on the events that marked the evolutionary history of this operon, both in terms of its gene content and in terms of its regulatory sequence.
Experimental procedures
Bacterial strains and growth conditions
Strains of bacterial species used were P. putida KT2440, P. aeruginosa PAO1, A. tumefaciens C58, S. meliloti 1021 and X. campestris ATCC33913. All of them were grown in their respective suitable growth and temperature conditions as recommended (Miller, 1991). A P. putida lexA2(Def) mutant was constructed by marker exchange with a lexA2 gene interrupted by the insertion of a ΩKm cassette in a SmaI restriction site generated by inverse PCR in its +301 nucleotide, following previously described methodology (Campoy et al., 2003) and using a pGP704 suicide vector-derivative (pUA658) carrying gentamicin resistance (Fernández de Henestrosa et al., 1997). Antibiotic concentrations used for each microorganism were as previously reported (Miller, 1991).
Nucleic acids techniques
RNA and DNA total extraction was carried out by standard methods (Sambrook and Russell, 2001). Genes and promoter fragments for electrophoretic mobility shift assays were isolated by PCR from total DNA extraction of each bacterial species, using suitable oligonucleotide primers designed in accordance to their published sequences. Directed mutagenesis of the P. putida lexA2 promoter was carried out by PCR as described earlier (Campoy et al., 2003) and the presence of DNA changes was confirmed by sequencing using the dideoxy method (Sanger et al., 1977) on an ALF Sequencer (Pharmacia Biotech). RT-PCR analysis of gene expression was performed for all bacterial species as reported (Campoy et al., 2003) and using specific internal oligonucleotide primers for each gene. For each bacterial species (A. tumefaciens, P. putida, P. aeruginosa, S. meliloti and X. campestris), the RNA concentration of the gene to be analysed was always normalized to that of its own trpA gene, because expression of the latter is not affected by DNA damage (Courcelle et al., 2001).
Purification of LexA proteins
LexA protein from Anabaena PCC7120 had been purified previously (Mazón et al., 2004). The lexA1 and lexA2 genes from P. putida, as well those of X. campestris and S. meliloti, were cloned by PCR using specific primers for each one. The 5′ end of the upper primer in all cases contained an NdeI restriction site in which the ATG initial triplet of each lexA gene to be cloned was included. The lower primer always started 200 bp downstream of the translational stop codon of the lexA genes. PCR fragments containing the desired lexA gene were cloned into a pGEM-T vector and, afterwards, inserted into a pET15b expression vector. pET15b-derivatives containing the appropriate lexA gene were transformed into the E. coli lexA (Def) BL21(DE3) codon plus strain (Fernández de Henestrosa et al., 2000) for overexpression of the corresponding LexA proteins, which were subsequently purified using the TalonTM Metal Affinity Resin Kit (Clontech) as reported (Mazón et al., 2004). All LexA proteins obtained were above 95% purity as determined with Coomassie Blue staining of SDS-PAGE (15%) polyacrylamide gels (data not shown) following standard methodology (Laemmli, 1970).
Electrophoresis mobility shift assays
LexA-DNA binding was analysed for each bacterial species by EMSAs using purified LexA proteins. DNA probes were prepared by PCR amplification with one of the primers labelled at its 5′ end with digoxigenin (DIG) and purifying each product in a 2–3% low-melting-point agarose gel. DNA-protein reactions (20 µl) typically containing 20 ng of the DIG-DNA-labelled probe and 80 nM of purified LexA protein were incubated in binding buffer: 10 mM N-2-Hydroxyethyl-piperazine-N ′ 2-ethanesulphonic acid (Hepes), NaOH (pH 8), 10 mM Tris-HCl (pH 8), 5% glycerol, 50 mM KCl, 1 mM EDTA, 1 mM DTT, 1 mg ml−1 salmon DNA and 50 µg ml−1 BSA. After 30 min at 30°C, the mixture was loaded onto a 6% non-denaturing Tris-glycine polyacrylamide gel [pre-run for 30 min at 10 V cm−1 in 25 mM Tris-HCl (pH 8.5), 250 mM glycine, 1 mM EDTA]. DNA-protein complexes were separated at 150 V for 60 min, followed by transfer to a Biodine B nylon membrane (Pall Gelman Laboratory). DIG-labelled DNA-protein complexes were detected following the manufacturer protocol (Roche). For the binding-competition experiments, a 300-fold molar excess of either specific or non-specific unlabelled competitor DNA was also included in the mixture. All EMSAs were repeated a minimum of three times to ensure reproducibility of results.
In silico phylogenetic analysis
Protein sequences for each gene under study were aligned using the clustalw program (Higgins et al., 1994). Multiple alignments were then used to infer phylogenetic trees with the seqboot, proml and consense programs of the Phylip 3.6 software package (Felsenstein, 1989), applying the maximum-likelihood method on 100 bootstrap replicates. The resulting phylogeny trees were plotted using TreeView (Page, 1996).
Acknowledgements
This work was funded by Grants BMC2001-2065 from the Ministerio de Ciencia y Tecnología (MCyT) de España and 2001SGR-206 from the Departament d’Universitats, Recerca i Societat de la Informació (DURSI) de la Generalitat de Catalunya, and by the Consejo Superior de Investigaciones Científicas (CSIC). M. Abella and M. Jara were recipients of predoctoral fellowships from the Ministerio de Educación y Cultura (MEC) and the DURSI respectively. We are deeply indebted to Joan Ruiz and Dr Pilar Cortés for their excellent technical assistance.




