RETRACTED: Deciphering the role of wound healing genes in skin cutaneous melanoma: Insights into expression, methylation, mutations, and therapeutic implications
Abstract
Skin Cutaneous Melanoma (SKCM) is a form of cancer that originates in the pigment-producing cells, known as melanocytes, of the skin. Delay wound healing is often correlated with the occurrence of and progression of SKCM. In this comprehensive study, we investigated the intricate roles of two important wound healing genes in SKCM, including Matrix Metalloproteinase-2 (MMP2) and Matrix Metalloproteinase-9 (MMP9). Through a multi-faceted approach, we collected clinical samples, conducted molecular experiments, including RT-qPCR, bisulphite sequencing, cell culture, cell Counting Kit-8, colony formation, and wound healing assays. Beside this, we also used various other databases/tools/approaches for additional analysis including, UALCAN, GEPIA, HPA, MEXPRESS, cBioPortal, KM plotter, DrugBank, and molecular docking. Our results revealed a significant up-regulation of MMP2 and MMP9 in SKCM tissues compared to normal counterparts. Moreover, promoter methylation analysis suggested an epigenetic regulatory mechanism. Validations using TCGA datasets and immunohistochemistry emphasized the clinical relevance of MMP2 and MMP9 dysregulation. Functional assays demonstrated their synergistic impact on proliferation and migration in SKCM cells. Furthermore, we identified potential therapeutic candidates, Estradiol and Calcitriol, through drug prediction and molecular docking analyses. These compounds exhibited binding affinities, suggesting their potential as MMP2/MMP9 inhibitors. Overall, our study elucidates the diagnostic, prognostic, and therapeutic implications of MMP2 and MMP9 in SKCM, shedding light on their complex interplay in SKCM occurrence and progression.
1 INTRODUCTION
Skin cutaneous melanoma (SKCM) stands out as the most aggressive form of skin cancer, fuelled by a rising global incidence that necessitates a profound exploration of its molecular intricacies.1, 2 SKCM is primarily caused by exposure to ultraviolet (UV) radiation from sunlight, making fair-skinned individuals more vulnerable. Genetic factors, including mutations in genes like BRAF and NRAS, contribute to susceptibility.3, 4 UV-induced DNA damage and immune responses also play pivotal roles in the initiation and progression of this aggressive form of skin cancer.5 Among the pivotal contributors to SKCM pathogenesis are Matrix Metalloproteinase family members, particularly Matrix Metalloproteinase 2 (MMP2) and Matrix Metalloproteinase 9 (MMP9), known for their critical roles in tumour development and metastasis by delaying wound healing.6, 7
Existing studies have extensively explored the multifaceted roles of MMP2 and MMP9 in wound healing and cancer progression.6, 8 While direct evidence linking MMP2 and MMP9 to cancer by delaying wound healing is limited, foundational research has investigated their functions in tissue repair and angiogenesis, processes integral to both wound healing and tumour development.9, 10 Studies have elucidated MMP2 and MMP9 influence on cell proliferation and morphogenesis in cancer cells, indirectly suggesting potential implications for wound healing dynamics.11, 12 Although the explicit connection remains to be firmly established, these investigations emphasized the intricate interplay between MMP2 and MMP9, wound healing, and cancer, warranting further research to unveil the underlying mechanisms and therapeutic implications.
The present study was designed to unravel the diagnostic and prognostic implications of MMP2 and MMP9 in SKCM. Additionally, it seeks to explore the potential oncogenic roles of these genes by investigating their impact on delaying wound healing in SKCM. For this purpose, this research employs a dual approach, integrating in silico analyses with molecular experiments, to comprehensively assess the functions of MMP2 and MMP9 in SKCM. By examining their diagnostic and prognostic roles and exploring their potential contribution to SKCM progression through delayed wound healing, the study aims to provide a comprehensive understanding of the molecular landscape of SKCM and potentially uncover novel therapeutic targets.
2 METHODOLOGY
2.1 Sample collection
We collected a total of 40 SKCM and 40 paired control samples from Nishtar Hospital in Multan, Pakistan, spanning from January 2021 to June 2023. This research received ethical approval from the Research Ethics Committee of PRAC, Pakistan. The sample collection adhered strictly to the Helsinki guidelines,13 and explicit informed consent was secured from each patient before sample collection process.
2.2 Nucleic acid extraction
DNA extraction employed an organic method,14 while RNA isolation utilized the TRIzol method.15 Subsequently, the purity of both DNA and RNA was assessed through the 280/260 ratio method, ensuring the integrity and quality of the nucleic acid samples.
2.3 RT-qPCR analysis
- GAPDH-F 5′-ACCCACTCCTCCACCTTTGAC-3′.
- GAPDH-R 5′-CTGTTGCTGTAGCCAAATTCG-3′.
- MMP2-F: 5′-AGCGAGTGGATGCCGCCTTTAA-3′.
- MMP2-R: 5′-CATTCCAGGCATCTGCGATGAG-3′.
- MMP9-F: 5′-GCCACTACTGTGCCTTTGAGTC-3′.
- MMP9-R: 5′-CCCTCAGAGAATCGCCAGTACT-3′.
3 BISULPHITE SEQUENCING ANALYSIS
3.1 Library preparation for targeted bisulphite sequencing analysis
First step involved the fragmentation of 1 μg of total DNA into fragments of approximately 200–300 bp using the Covarias sonication system (Covarias, Woburn, MA, USA). Subsequently, the DNA fragments underwent a series of processes, including repair and phosphorylation of blunt ends, facilitated by a mixture of enzymes such as T4 DNA polymerase, Klenow Fragment, and T4 polynucleotide kinase. The repaired fragments were then subject to 3′ adenylation using Klenow Fragment (3′-5′ exo-), followed by ligation with adapters. These adapters contained 5′-methylcytosine in place of 5′-cytosine and index sequences, and the ligation was executed using T4 DNA Ligase. After the construction of libraries, quantification was performed utilizing a Qubit fluorometer with the Quant-iT dsDNA HS Assay Kit (Invitrogen, Carlsbad, CA, USA). The prepared libraries were subsequently dispatched to Beijing Genomic Institute (BGI), China, for the purpose of targeted bisulphite sequencing. Once the sequencing was completed, the methylation data was subjected to a normalization process, resulting in the generation of beta values.
3.2 Receiver operating characteristic (ROC)
ROC curves based on expression and promoter methylation were employed to assess the potential of MMP2 and MMP9 as diagnostic biomarkers for detecting SKCM.
3.3 Validation of MMP2 and MMP9 expression using additional SKCM cohorts
OncoDB and GEPIA are invaluable tools for cancer research. OncoDB provides user-friendly access to The Cancer Genome Atlas (TCGA) data, facilitating gene expression analyses across various cancer types.16 GEPIA, on the other hand, offers interactive visualization and analysis of RNA sequencing expression data from diverse cancer databases, aiding in the exploration of gene functions and pathways.17 In the present study, both these tools were used for the mRNA expression validation of MMP2 and MMP9 in TCGA datasets.
The Human Protein Atlas (HPA) is a comprehensive resource providing information on the human proteome.18 It offers detailed protein expression profiles in various tissues and organs, aiding researchers in understanding protein localization, abundance, and potential roles in health and disease. In the current study, HPA was used to validate the protein expression of MMP2 and MMP9 in SKCM tissue.
3.4 Promoter methylation validation of MMP2 and MMP9 using additional SKCM cohorts
MEXPRESS is an essential database for exploring DNA methylation patterns across diverse cancer types.16 Offering an intuitive interface, it enables researchers to analyse and visualize relationships between gene expression and methylation, providing valuable insights into the molecular mechanisms underlying cancer development and progression. In the current research, MEXPRESS was used to validate promoter methylation levels of MMP2 and MMP9 in TCGA SKCM dataset.
3.5 Mutational analysis of MMP2 and MMP9
cBioPortal is a powerful platform for exploring cancer genomics data.19 It facilitates the visualization and analysis of complex genetic alterations in various cancers, enabling researchers to interpret genomic profiles, identify potential therapeutic targets, and enhance understanding of the molecular basis of cancer, ultimately contributing to precision medicine advancements. In this work, cBioPortal database was utilized for the mutational analysis of MMP2 and MMP9 in TCGA SKCM dataset.
3.6 Prognostic model development
KM Plotter is a valuable online tool for survival analysis in cancer research.20 It allows users to assess the impact of specific genes on patient survival across multiple cancer types. In our work we used this resource for the survival analysis of MMP2 and MMP9 in across SKCM patients.
The least absolute shrinkage and selection operator (Lasso) and multivariate Cox proportional hazard regression analysis were further developed to construct the prediction model with the ‘survival’ package in R language.21 In this analysis, the TCGA-SKCM dataset was used as the training set, and GSE100797, GSE133713, GSE190113, and GSE59455 datasets were used as the validation datasets. In this analysis, positive coefficients indicate increased risk of an event (e.g. death), while negative coefficients suggest reduced risk. Their magnitude signifies the impact of variables on hazard rates, aiding in building prognostic models for survival outcomes. The formula of the prognostic model of SKCM patients' prognosis was as follows: risk score = the sum of the multivariate Cox regression coefficient variation of each mRNA.
3.7 Gene enrichment analysis of MMP2 and MMP9
DAVID (Database for Annotation, Visualization, and Integrated Discovery) is a bioinformatics resource offering comprehensive functional annotation tools for researchers.22 It aids in understanding biological meaning behind large gene lists by uncovering enriched biological themes. In this work, DAVID was used to perform gene enrichment analysis of MMP2 and MMP9.
3.8 Cell culture
The A2058 cell line, derived from human malignant melanoma, was obtained from Pricella (Wuhan, Hubei, China), and its authenticity was confirmed through STR matching analysis. The A2058 cells were cultured under standard conditions at 37°C with 95% humidity and 5% CO2 to maintain their viability and characteristics.
3.9 Knockdown of MMP2 and MMP9 in SKCM cell line
SiRNAs targeting MMP2 and MMP9 were acquired from OBiO Company. For MMP2 and MMP9 knockdown, melanoma cell lines underwent transfection with siRNA using a Transfection Reagent (INTERFERin, French).
3.10 Cell Counting Kit-8 and colony formation assays
The cell proliferation ability of melanoma cells was evaluated utilizing the Cell Counting Kit-8 (CCK-8, APExBIO, USA). Briefly, 3 × 103 cells per well were seeded into 96-well plates 24 h post-transfection. After incubation at 37°C for various time points (0, 24, 48, and 72 h), CCK-8 reagent was introduced to each well, and the absorbance was measured at 450 nm. For the colony formation assay, 5 × 102 melanoma cells were cultured in 6-well plates for 10 days under 37°C and 5% CO2 conditions. Subsequently, the cells were stained with 0.1% crystal violet for 15 min, and colony quantification was performed using ImageJ software.
3.11 Wound healing assay
Following transfection, A2058 cells were allowed to reach approximately 80% confluence when cultured in 6-well plates. At this point, a linear scratch was created using a pipette. Subsequently, cell images were captured at both 0 and 24 h post-wounding. The assessment of cell migration was performed using ImageJ software by applying the formula: cell migration index = (scratch area at 0 h − scratch area at 24 h)/scratch area at 0 h.
4 DRUGS ACQUISITION AND MOLECULAR DOCKING ANALYSIS
4.1 Drugs acquisition
DrugBank is a comprehensive pharmacological resource providing detailed information on drugs and drug targets.23 It amalgamates data on drug mechanisms, interactions, and adverse effects, aiding researchers in drug discovery and development. In the present study, DrugBank database was used to predict MMP2 and MMP9 inhibitory drugs.
4.2 Ligand and receptor preparation and docking analysis
To assess the binding affinities between two chosen drugs, Estradiol and Calcitriol, and the MMP2 and MMP9 proteins, we employed a molecular docking approach facilitated by the CB-DOCK web server (http://clab.labshare.cn/cb-dock/php/).24 We obtained the SDF structures of Estradiol and Calcitriol from the PubChem database, and for the MMP2 and MM9 proteins, we generated PDB structures using the SwissModel tool. The process involved several key steps, including ligand (Estradiol and Calcitriol) pre-processing, the removal of excess ligands from the target proteins (macromolecules), elimination of crystal water molecules, and the addition of hydrogen atoms. The molecular docking was carried out using the CB-DOCK platform to calculate the binding energies of the molecules across various conformations. The conformation with the highest hydrogen bond energy was selected as the active component of the protein interaction. For visualization purposes, we employed PYMOL software (version 2.5.2).
4.3 Statistics
In this study, for GO and KEGG enrichment analysis, we employed the Fisher's Exact test to compute the differences.25 Correlational analyses were carried out using Pearson method. A Student t-test was employed for making comparisons. All the analyses were conducted using R version 3.6.3 software.
5 RESULTS
5.1 RT-qPCR analysis of MMP2 and MMP9 in clinical samples
Employing RT-qPCR, we assessed the expression levels of MMP2 and MMP9 in 40 paired SKCM tissue and corresponding normal tissue samples. Statistical analysis using SPSS software revealed a significant up-regulation of MMP2 and MMP9 in SKCM samples compared to their non-cancerous counterparts (p-value <0.05) (Figure 1A). Upon further analyses to assess MMP2 and MMP9 as potential SKCM biomarkers, the area under the ROC curve surpassed 0.9, while both sensitivity and specificity exceeded 97% (Figure 2B). Based on the expression data, these findings emphasized the promising potential of MMP2 and MMP9 as effective biomarkers for this malignancy.
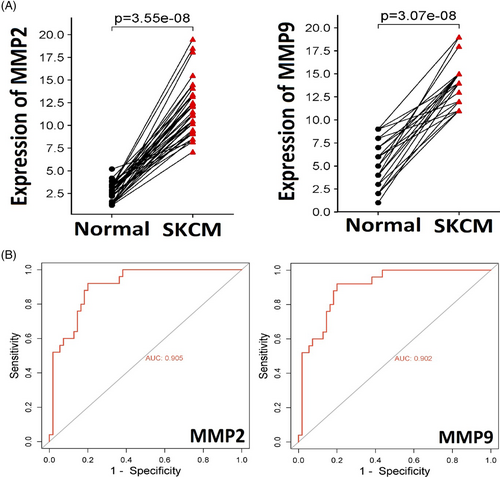
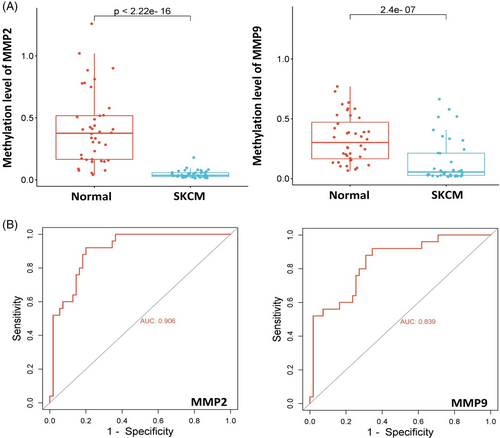
5.2 Promoter methylation analysis of MMP2 and MMP9 in clinical samples
Dysregulation of the MMP2 and MMP9 can be due to aberrant DNA methylation. Therefore, utilizing the bisulphite sequencing method, we examined the promoter methylation levels of MMP2 and MMP9 in normal (n = 40) and SKCM (n = 40) tissue samples. Our analysis revealed a significant decrease (p-value <0.05) in MMP2 and MMP9 promoter methylation levels in the SKCM sample group compared to the normal sample group (Figure 2A). The methylation data exhibited an area under the ROC curve exceeding 0.8, with both sensitivity and specificity surpassing 97% (Figure 2B). These results further highlight the potential of MMP2 and MMP9 methylation levels as valuable biomarkers in SKCM.
5.3 Validation of MMP2 and MMP9 expression using additional SKCM cohorts
In this phase of our investigation, we initially validated the mRNA expression of MMP2 and MMP9 in normal and SKCM specimens using TCGA datasets through the OncoDB and GEPIA web tools. Our findings indicated a significant (p-value <0.05) elevation in MMP2 and MMP9 mRNA levels in the SKCM tissue sample groups compared to the normal tissue sample group (Figure 3A,B).
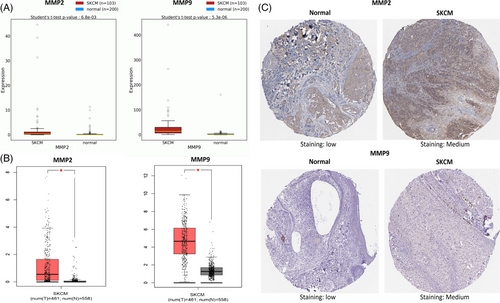
Next, the HPA database was utilized to validate the immunohistochemistry-based protein expression of MMP2 and MMP9 in SKCM and normal tissues samples. Immunohistochemistry images showed that the protein expression of MMP2 and MMP9 was relatively high (Staining: medium) in SKCM tissue samples as compare to the normal samples having low expression of these proteins (Staining: low) (Figure 3C).
5.4 Promoter methylation validation of MMP2 and MMP9 using additional SKCM cohorts
To further validate the promoter methylation levels of MMP2 and MMP9 genes in SKCM, we utilized the MEXPRESS database. The results from MEXPRESS corroborated a negative correlation (p-value <0.05) between mRNA expression and promoter methylation levels of MMP2 and MMP9 in SKCM (Figure 4). This additional confirmation supports the notion that specific promoter methylation sites may contribute to the up-regulation of MMP2 and MMP9 expression.
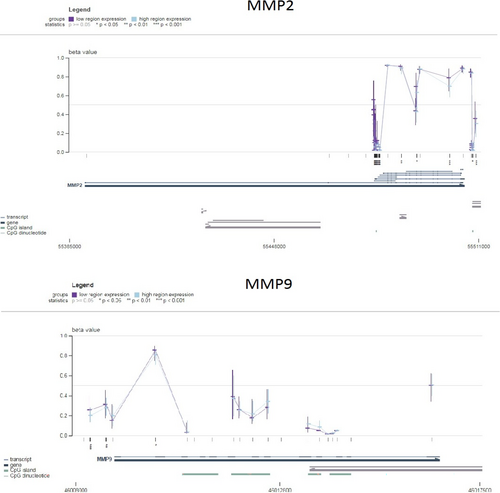
5.5 Mutational analysis of MMP2 and MMP9
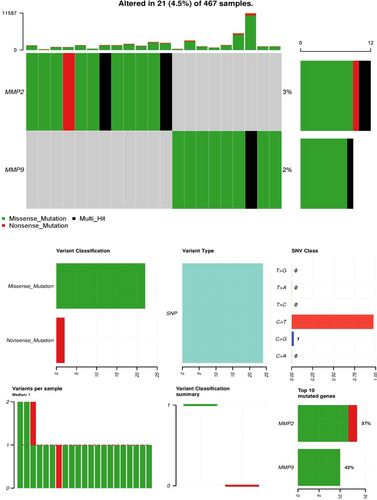
To comprehend the mutational profile of MMP2 and MMP9 genes in SKCM, we examined their genetic alterations using cBioPortal. The mutation rates for MMP2 and MMP9 were determined to be 3% and 2%, respectively, among the analysed SKCM samples (Figure 5). Additionally, the C > T genetic mutation was the most prevalent type of mutations in MMP2 and MMP9 genes across SKCM samples (Figure 5). However, given the low frequencies of MMP2 and MMP9 mutations, it is speculated that the substantial up-regulation of MMP2 and MMP9 expression in SKCM specimens may not be attributed to these mutations.
5.6 Prognostic model development
The prognostic relevance of MMP2 and MMP9 expression in SKCM patients was evaluated using the KM Plotter tool. Higher expression levels of MMP2 and MMP9 were closely associated with poor overall survival in SKCM patients (Figure 6A).
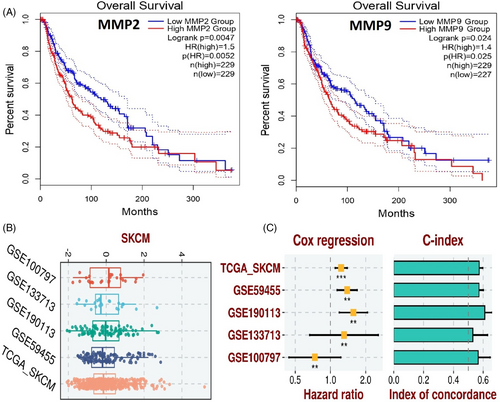
For developing the prognostic model based on MMP2 and MMP9 genes, we utilized the TCGA-SKCM dataset as the training set, and GSE100797, GSE133713, GSE190113, and GSE59455 datasets as the validation sets. The construction of the prognostic model involved a stepwise Cox regression method, considering parameters like hazard ratio, c-index, and risk score. The evaluation of our predictive prognostic model using the c-index revealed its effective and robust performance in assessing the prognosis of SKCM patients across all analysed datasets (Figure 6B,C).
5.7 Gene enrichment analysis
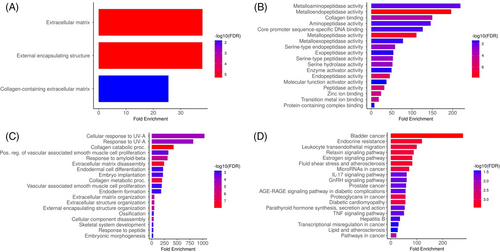
We analysed hub genes to figure out their GO and KEGG pathways in SKCM. In the CC, ‘Extracellular matrix, external encapsulating structure, and collagen-containing extracellular matrix’ etc., terms were significantly associated with the MMP2 and MMP9 (Figure 7A). Concerning MF, the ‘Metalloaminopeptidase activity, Metallendopeptidase activity, and collagen binding’ etc., terms were closely associated with the MMP2 and MMP9 (Figure 7B). In BP, some vital functions including ‘Cellular response to UV-A, response to UV-A, and collagen catabolic proc’ etc., terms were significantly associated with the MMP2 and MMP9 (Figure 7C). Moreover, MMP2 and MMP9-enriched KEGG pathways include ‘Bladder cancer, endocrine resistance, and relaxin signalling pathway’ etc., (Figure 7D).
5.8 Functional verification of the in vitro and in vivo roles of MMP2 and MMP9 in SKCM
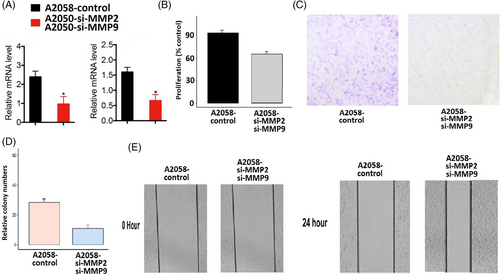
The MMP2 and MMP9 genes work synergistically to regulate processes such as tissue remodelling, wound healing, and cancer invasion. Therefore, the simultaneous silencing of MMP2 and MMP9 and was carried out in A2058 cells using siRNA to analyse their functional synergetic impact on the different parameters. Silencing efficiency was confirmed by RT-qPCR, revealing reduced expression of MMP2 and MMP9 in transfected A2058 cells compared to controls (Figure 8A). Subsequent analyses, including CCK-8 and colony-forming assays, demonstrated that MMP2 and MMP9 knockdown led to decreased cellular proliferation relative to control A2058 cells (Figure 8B,D). Moreover, diminished expression of these genes correlated with increased migratory capabilities in these cells (Figure 8E).
5.9 Drug prediction and molecular docking analysis
A search in the DrugBank database was conducted to identify potential drugs capable of down-regulating the expression of MMP2 and MMP9 genes in the context of SKCM treatment. The results revealed two promising drugs, namely Estradiol and Calcitriol, within this database, showing the potential to decrease the expression of MMP2 and MMP9 genes.
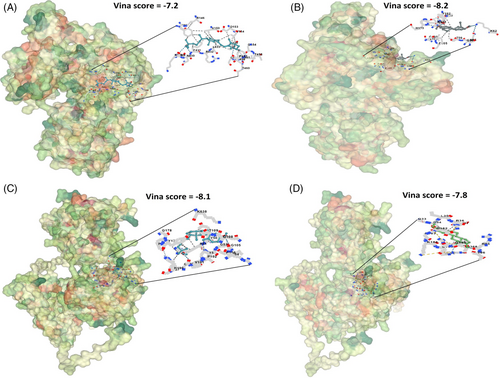
In the subsequent phase, we further validated the impact of Estradiol and Calcitriol on the reduction of expression through molecular docking analysis. Docking results exhibited varying binding affinities of Estradiol and Calcitriol with MMP2 and MMP9, ranging from −7.2 to −8.2 kcal/mol (Figure 9). These binding affinities, within the range of −7.2 to −8.2 kcal/mol, indicate a relatively robust interaction between Estradiol and Calcitriol with MMP2 and MMP9 proteins (Figure 9). To ascertain their actual efficacy as inhibitors for MMP2 and MMP9 proteins, comprehensive in vitro and in vivo studies would be necessary, despite the indicative strong binding affinities.
6 DISCUSSION
Our investigation into the roles of two important wound healing genes, including MMP2 and MMP9 in SKCM has provided a comprehensive understanding of their diagnostic, prognostic, therapeutic, and functional implications in this malignancy. The up-regulation of MMP2 and MMP9 in SKCM tissues, as consistently demonstrated across clinical samples from the local SKCM patients, suggests their potential as robust diagnostic biomarkers. In physiological wound healing, MMPs play a crucial role in tissue remodelling, allowing for controlled cell migration and matrix degradation.26, 27 However, in SKCM, the dysregulated and overactive MMP2 and MMP9 may lead to excessive matrix degradation, impairing the orderly healing process. This disruption can result in delayed wound healing, as the balance between tissue degradation and regeneration is disturbed.28, 29 The persistent activity of MMP2 and MMP9 may further promote tumour invasion, angiogenesis, and immune evasion, fostering an environment conducive to SKCM development and progression.30, 31 This aligns with previous studies highlighting the dysregulation of MMP2 and MMP9 in various cancers, such breast cancer, lung cancer, and colorectal cancer, emphasizing their diagnostic relevance.32-34 The validation of our findings across diverse datasets, including TCGA and the HPA, strengthens the generalizability of our results and emphasize the clinical significance of MMP2 and MMP9 in SKCM diagnosis.
The observed decrease in promoter methylation levels of MMP2 and MMP9 in clinical SKCM tissues samples taken from the local patients implies a potential epigenetic regulatory mechanism contributing to their overexpression. Aberrant DNA methylation is a well-established mechanism in cancer progression, impacting gene expression patterns.35, 36 Our findings align with studies suggesting that promoter hypomethylation contributes to the overexpression of MMPs in cancer, facilitating tumour invasion and metastasis.37, 38 The negative correlation between promoter methylation and mRNA expression, validated through MEXPRESS, further substantiates the regulatory impact of DNA methylation on MMP2 and MMP9 in SKCM.
The analysis of mutational landscapes revealed low mutation rates for MMP2 and MMP9, suggesting that genetic alterations may not be the primary drivers of their dysregulation in SKCM. This is consistent with the notion that epigenetic modifications and other regulatory mechanisms, such as microRNA-mediated control, could play pivotal roles in MMP2 and MMP9 expression in cancer.39 The prognostic significance of MMP2 and MMP9, as evidenced by KM Plotter analysis, aligns with studies indicating their association with adverse clinical outcomes in various cancers, including breast, colorectal, and lung cancers.40-42 Furthermore, the development of a prognostic model based on MMP2 and MMP9 genes, utilizing both training and validation datasets, adds a predictive dimension to their clinical relevance. The robustness of the model, demonstrated by high c-index values across diverse datasets, underscores its potential utility in assessing SKCM patient prognosis. Prognostic models incorporating molecular markers are increasingly recognized as valuable tools for personalized medicine and treatment stratification in cancer.43, 44
The gene enrichment analysis highlighted the involvement of MMP2 and MMP9 in processes related to extracellular matrix organization, response to UV-A, and various collagen-related functions. These findings align with the established roles of MMPs in tumour microenvironment remodelling, immune evasion, and cancer cell migration.45, 46 The KEGG pathway analysis implicating MMP2 and MMP9 in pathways such as bladder cancer and endocrine resistance provides insights into potential therapeutic targets within these pathways.
Functional verification experiments, including knockdown assays, confirmed the synergistic impact of MMP2 and MMP9 on cellular proliferation and migration in A2058 cells. These results align with the recognized roles of MMPs in promoting tumour progression and metastasis in other cancer types.47, 48 The identification of potential therapeutic drugs, Estradiol and Calcitriol, through the DrugBank database, opens avenues for targeted interventions. Estradiol, an oestrogen derivative, displays anti-proliferative effects and can modulate hormone receptors, offering a potential avenue for hormone-responsive cancers.49 Calcitriol, the active form of vitamin D, possesses anti-inflammatory and immune-modulatory properties, contributing to its anti-cancer potential.50 Molecular docking analyses suggest interactions between Estradiol, Calcitriol, and MMP2/MMP9 proteins, indicating a potential inhibitory impact on the MMP2 and MMP9. However, their clinical efficacy in SKCM necessitates rigorous validation through comprehensive in vitro, in vivo, and clinical studies.
7 CONCLUSION
In conclusion, our study revealed the intricate involvement of MMP2 and MMP9 in SKCM, shedding light on their diagnostic and prognostic potentials, therapeutic significance, and functional impact on SKCM progression by delaying wound healing process. The integration of clinical data, in silico validations, drug prediction, and molecular docking analyses enhances the translational relevance of our findings. Overall, the findings of this study offer promising avenues for further research and potential clinical application of MMP2 and MMP9 in SKCM management.
ACKNOWLEDGEMENTS
The authors would like to extend their sincere appreciation to the Researchers Supporting Project Number (RSPD2024R986), King Saud University, Riyadh, Saudi Arabia.
CONFLICT OF INTEREST STATEMENT
The authors declare that there is no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The authors would like to provide the data if required.




