LncRNA00492 is required for marginal zone B-cell development
Funding Information
This study was supported by the Graduate Research and Innovation Projects of Jiangsu Province (KYCX19_0114).
Abstract
B-cell development undergoes a series of steps from the bone marrow to the secondary lymphoid organs. A defect in B-cell development can lead to immunodeficiency or malignant disorders, such as leukaemia or lymphoma. Long non-coding RNAs have been reported to act as important regulators of many pathological processes. However, very little is known regarding the role of lncRNAs during B-cell development and the regulation of their expression. In this study, we explored the expression and role of lncRNA Gme00492 in B-cell development. We observed that lnc00492 was highly expressed in B-cell development and primarily expressed in the nucleus. Lnc00492-deficient mice had fewer marginal zone B cells in the spleen, likely due to a developmental block. Importantly, lnc00492 interacts with CTBP1 and targets it for ubiquitination and degradation during B-cell development, whereas the transcriptional corepressor factor CTBP1 plays a critical role in Notch2 signalling. Thus, we identified a novel regulatory axis between lnc00492 and CTBP1 in B cells, suggesting that lnc00492 is essential for marginal zone B-cell development.
Abbreviations
-
- BM
-
- bone marrow
-
- CHX
-
- cycloheximide
-
- CPAT
-
- coding potential assessment tool
-
- CTBP1
-
- C-terminal binding proteins-1
-
- FACS
-
- fluorescence-activated cell sorting
-
- FISH
-
- fluorescence in situ hybridization assay
-
- FO
-
- follicular
-
- Hes1
-
- hairy and enhancer of split homolog-1
-
- Hes5
-
- hairy and enhancer of split homolog-5
-
- LN
-
- lymph nodes
-
- LncRNAs
-
- long non-coding RNAs
-
- MZ
-
- marginal zone
-
- qRT-PCR
-
- quantitative real-time PCR
-
- RIP
-
- RNA-binding protein immunoprecipitation assays
INTRODUCTION
B cells are the central elements of the adaptive immune system and protect against a nearly unlimited variety of pathogens. [1 Defects in B-cell development and function lead to autoimmunity, malignancy and immunodeficiencies. [2 B-cell development undergoes a series of steps from the bone marrow to the secondary lymphoid organs. [3 In bone marrow, B cells develop from a lymphoid precursor and then transit through the pro-B cell, pre-B cell and immature B-cell stages. [4 Immature B cells travel to the spleen and complete maturation. In the process, those B cells include T1 and T2 B-cell subsets. In the spleen, there are follicular (FO) B cells and marginal zone (MZ) B cells expressing immunoglobulin. [5, 6 The FO B-cell subset plays an important role in T-dependent B-cell immune responses. [7 The MZ B-cell subset is critical for T-independent B-cell immune responses. [8 Many signalling pathways participate in the regulation of transitional subsets that develop into MZ B cells, including Notch2 signalling, BCR signalling and NF-κB signalling. [9 Among them, the most important pathway is the Notch signalling pathway. Any element changes in the Notch pathway will result in abnormal development of MZ B cells. [10
Long noncoding RNAs (lncRNAs) are non-protein-coding RNA, which is greater than 200 nt in length. [11 Many studies have implicated lncRNAs in a wide range of biological processes, including immune function, [12 autoimmunity [13 and oncogenesis. [14 To date, most lncRNAs are expressed in the cytoplasm and function as ceRNAs interacting with miRNAs or as binding RNAs interacting with proteins. [15 Other lncRNAs are expressed in the nucleus and function as guides, decoys or scaffolds. [16 Recent work has shown that lncRNAs have emerging roles in innate and adaptive immunity. For example, in T cells, lncRNA AW112010 promotes the differentiation of T cells by suppressing IL-10 expression through histone demethylation. [17 In monocytes, IL1b-eRNA is required for LPS-induced proinflammatory responses. [18 In dendritic cells, lnc-DCs are required for normal differentiation and function. [19, 20 Thus, lncRNAs play a critical role in the differentiation and maturation of immune T cells, NK cells and macrophages. However, the biological functions and underlying mechanisms of lncRNAs in B cells remain largely unknown.
In this study, we found that the expression level of lnc00492 was positively correlated with the maturation of B cells by screening the high-throughput sequencing results. We confirmed that lnc00492 is a novel long non-coding RNA and that there are no reports about lnc00492 biological functions in B cells. Therefore, understanding the function and molecular mechanism of lnc00492 in B-cell development may provide a reference for the treatment of autoimmune diseases and B cell-related tumours in clinic.
MATERIALS AND METHODS
Mice
Lnc00492± mice were purchased from GemPharmatech Co., Ltd., Jiangsu, China. lnc00492−/− and lnc00492+/+ mice were obtained by breeding two lnc00492± mice. All the experiments were performed using mice between 8 and 12 weeks old. All the mice were bred and maintained in the Animal Center of the Southeast University. The animal study was approved by the Ethics Committee of Southeast University (20200701007, 01 July 2020).
Flow Cytometry
Bone marrow, spleen and LN were isolated from euthanized mice and homogenized to harvest total cells. The cells were re-suspended in 2 ml RBC lysis buffer and incubated for 3 min at 37℃ to remove the erythrocytes. Samples were washed with staining buffer (PBS, 1% FBS). All samples were incubated with fluorochrome-conjugated antibodies and were filtered immediately before analysis to remove any clumps. For Ki67 staining, cells were stained with Ki67 in 1×PBS buffer and incubated for 40 min at 4℃. For annexin V staining, cells were stained with FITC-conjugated annexin V and incubated for 40 min at 4℃. Flow cytometry was performed on an LSRII, and data were analysed using FlowJo software. Fluorochrome-conjugated antibodies and subset definitions are listed in tables S1 and S2.
Cell Sorting
Bone marrow and spleen pooled from three mice and homogenized to harvest cells. After purified with mouse B isolation kit (STEMCELL Technologies Inc.), all samples were stained according to FACS surface markers as noted in table S1. Cells were sorted into pro-B, pre-B, immature-B, mature B, T1, T2, FO and MZ cells via FACS Aria.
Subcellular fractionation
Subcellular fractionation assays were performed with the PARIS Kit (Sigma-Aldrich). According to the instructions, sorted B cells were divided into nuclear and cytoplasm apart. In the two parts, the mRNA level of lnc00492 was examined by qRT-PCR. XIST and GAPDH were served as positive controls.
Fluorescence in situ hybridization assay (FISH)
FISH assays were performed essentially as described previously. [21 Briefly, probes for lnc00492 were synthesized with rhodamine modification (Sigma-Aldrich). After immobilized with 4% paraformaldehyde, the cells were treated with Triton X-100 (1%) and then mix the probes and cells for overnight at 37℃. In the last, DIPA was used to staining the nuclear of cells. Photo the cells with confocal microscope.
Quantitative real-time PCR (qRT-PCR)
TRIzol (Life Technologies, Carlsbad, U.S.A.) was used to extract total RNA. Cell lysates were incubated on ice for 10 min. Then, chloroform and isopropanol were added in turn. Finally, the RNA were solubled with DEPC water and reverse transcribed with kit (Cat#11119-11141; Yeasen, Shanghai, China). qRT-PCR was performed on ABI PRISM 7500 RT-PCR system (Applied Biosystems, Foster City, CA, U.S.A.) using SYBR Premix Ex Taq (Cat#11198-11200; Yeasen, Shanghai, China). The relative expression of target genes was calculated using the 2−ΔΔCT method. Primers are listed in table S3.
RNA pull-down mass spectrometry assays
RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to perform the experiment. Briefly, sense and anti-sense lnc00492 RNA were transcribed with a MEGAscript™ T7 Transcription Kit (Invitrogen, Carlsbad, CA, USA) and a Pierce RNA 3′End Desthiobiotinylation Kit (Thermo Fisher Scientific, Waltham, MA, USA). Lysates were prepared using lysis buffers. After being labelled with biotin, lncRNAs incubated with the cell lysis supernatant at 37℃ for 2 h. Then, the mixture of RNA-protein complexes was washed and eluted. The RNA–protein complexes were subjected to mass spectrometric analysis (Thermo Finnigan, San Jose, CA). We preformed LC-MS/MS experiments with an LTQ linear ion trap mass spectrometer equipped with a microspray source.
RNA-binding protein immunoprecipitation (RIP) assays
RNA-binding protein immunoprecipitation kit (Millipore, USA) was used to perform the RIP experiment. According to the instructions, the antibody was used to grab the binding RNA in B cells. After digested with proteinase K, the target RNA was extracted. qRT-PCR was preformed to detect the RNA level. The RNA levels were normalized to the input RNA levels (10%).
Cycloheximide treatment and Western blot
The stability of protein at indicated periods of time was detect with cycloheximide (100 μg/ml, Sigma). Briefly, B cells were incubated overnight at 37℃, 5% CO2. After washed with PBS, B cells were lysed with RIPA lysis buffer (Biosharp). The proteins were heated at 100℃ for 15 min. 10% SDS-PAGE gel and PVDF membranes (Beyotime Biotech Co., Ltd.) were used to separated total protein. After been blocked with 5% BSA, the membranes were incubated with corresponding primary antibodies at 4℃ for overnight. The antibodies are listed in table S1. ECL system (Millipore Biotech Co., Ltd.) was preformed to detect the protein bands.
Statistical analyses
All data were analysed with a two-tailed Student's t-test conducted using GraphPad Prism Software (GraphPad software, La Jolla, CA). Figures are graphed as the mean with the SD of the mean for continuous numerical data. Significance is shown as p < 0·05* and p < 0·01**.
RESULTS
Lnc00492 is upregulated in B-cell development
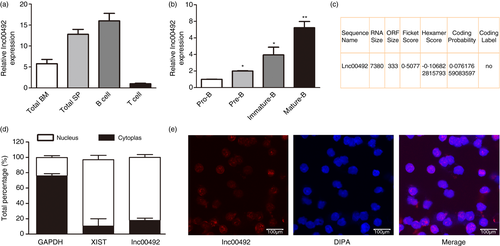
We first examined the expression of lnc00492 in mouse bone marrow and spleen cells as well as other types of immune cells. The results showed that lnc00492 expression was significantly higher in purified splenic B cells than in other immune cells and tissues. However, T cells expressed a low level of lnc00492 (Figure 1A). Further analysis of lnc00492 expression in sorted B-cell subset populations showed a positive correlation between lnc00492 expression and the degree of B-cell differentiation and maturation (Figure 1B). To confirm the non-coding potential of lnc00492, we preformed the online detecting with Coding Potential Assessment Tool (CPAT v2·0.0). [22 The value for lnc00492 obtained from the CPAT database indicates that lnc00492 is a noncoding RNA (Figure 1C). Additionally, we performed qRT-PCR to investigate the subcellular localization of lnc00492 and found that lnc00492 was primarily localized in the nuclear fraction of B cell (Figure 1D). To confirm the nuclear location of lnc00492, sorted B cells from mouse spleen were immediately fixed to perform lnc00492 RNA fluorescence in situ hybridization using labelled short oligo probes. The results also showed that lnc00492 was mainly expressed in the cell nucleus (Figure 1E). Altogether, these data suggest that lnc00492 plays a critical role in the regulation of B-cell development.
Lnc00492 deficiency impaired B-cell development
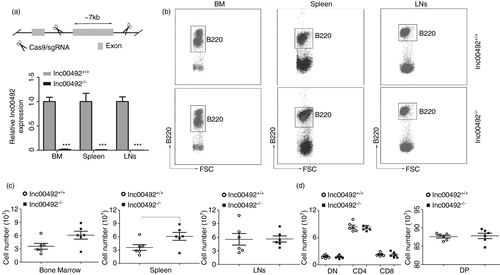
To understand the biological importance of lnc00492 up-regulation in B-cell development, we developed lnc00492 knockout mice with a CRISPR/Cas9-mediated strategy (Figure 2A). To confirm the depletion of lnc00492, we measured lnc00492 expression in lnc00492−/− and lnc00492+/+ mice by qRT-PCR. As shown in Figure 2A, lnc00492 was knocked out in B cells from the bone marrow (BM), spleen and lymph nodes (LN) of lnc00492−/− mice. To determine whether lnc00492 affects B-cell development, B220+ B cells were quantified in the BM, spleen and LN of lnc00492+/+ and lnc00492−/− mice. We found that lnc00492−/− mice had more total B cells in the spleen and no different B cells in the BM and LN (Figure 2B-C). In addition, CD4+ T lymphocyte and CD8+ T lymphocyte cell numbers did not change significantly in the thymus of lnc00492−/− mice (Figure 2D). These results indicate that lnc00492 may play an important role in spleen B-cell development.
Lnc00492 deficiency leads to impaired of MZ B-cell development in the spleen
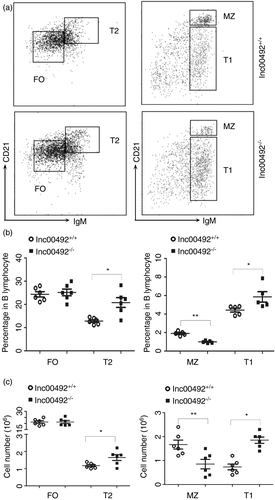
To better understand the function of lnc00492 in B cells, we detected the effect of lnc00492 on B-cell subsets in the spleen. According previous papers, We evaluated the proportion of T1 B cells, T2 B cells, FO B cells and MZ B cells from lnc00492+/+ and lnc00492−/− mice. The results indicated that compared with lnc0049+/+ mice, lnc00492−/− mice showed an increased frequency of T1 cells and T2 cells but a significant decrease in MZ B cells (Figure 3A). However, there were no differences in FO B cells from lnc00492+/+ and lnc00492−/− mice (Figure 3B). In addition, the overall number of T1 and T2 cells in lnc00492−/− mice was higher than that in lnc00492+/+ mice. In contrast, the MZ B-cell number presented a statistically significant decrease in lnc00492−/− mice. Lnc00492−/− mice revealed a comparable number of FO B cells (Figure 3C). Together, these results indicate that lnc00492 deficiency significantly inhibited the mature of MZ B-cell subset and increased the T1 and T2 B-cell subset in spleen. These phenomenon also explain that why lnc00492 is so effectively increased in normal B-cell development.
Decreased MZ B cells are not due to bone marrow development failure in lnc00492-deficient mice
To further explore the reasons of MZ B cells decreased in lnc00492−/− mice, we assessed the different of B-cell subset in upstream bone marrow. Our results showed that there were no significant differences in total survival (Figure 4A) and B220+ B cell (Figure 2B). Subsequently, we analysed B-cell precursor cells in the bone marrow of lnc00492+/+ and lnc00492−/− mice (Figure 4B). Lnc00492−/− mice showed a similar frequency and number of pro/pre-B, pro-B and pre-B cells (Figure 4C). Moreover, immature B cells and mature B cells were to be excluded comparable in lnc00492+/+ and lnc00492−/− mice, where the numbers were same (Figure 4D). Mature bone marrow B cells showed no difference in lnc00492−/− mice and lnc00492−/− mice. This may mean that there is no change in the recirculation score due to increased overall B-cell retention in the spleen.

Next, we further explored that whether the decrease of MZ B cells in the spleen from lnc00492−/− mice is related to retention cells reduced. However, our peripheral blood results indicated that there are no different in MZ B cells from lnc00492+/+ and lnc00492−/− mice (Figure 4E). Similarly, there were also shows same result in lymph nodes (Figure 4F). Furthermore, as we all know, after stimulated with antigens, MZ B-cell differentiate. Hence, we performed FACS analysis of macrophages and dendritic cells in the spleen to examine this possibility. The results showed that these populations were similar in lnc00492+/+ and lnc00492−/− mice (Figure 4G-H). However, the myeloid cells of lnc00492−/− mice were significantly increased, which is consistent with previous studies. [23, 24 Considering with no different in bone marrow development, these results indicate that the decreased of MZ B cells is due to the intrinsic defect of spleen.
Furthermore, we hypothesized that the decrease of MZ B cells in lnc00492−/− mice was related to the proliferation defect or apoptosis. We preformed the ki67 staining assays in B-cell subsets. Our results indicated that the number of Ki67-positive cells was similar in lnc00492+/+ and lnc00492−/− mouse MZ B cells (Figure 4I). Annexin V staining assays were preformed to detect the apoptosis of MZ B cells. The results also indicated there were no different between lnc00492+/+ and lnc00492−/− mice (Figure 4J). In summary, we concluded that the decrease of MZ B cells is because of the developmental block in T2 B cells not bone marrow development failure in lnc00492-deficient mice.
Lnc00492 physically interacts with CTBP1 in B cells
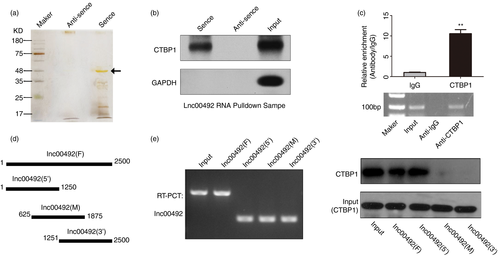
To explore the mechanism through which lnc00492 blocks MZ B-cell development in the spleen, we performed pull-down assays to identify the proteins interacting with lnc00492. Silver staining result indicates that there were a specific band in the sense lane close to 50 kDa (Figure 5A). Potential candidate lnc00492-interacting proteins were identified from three independent mass spectrometry experiments. We analyse the mass spectrometry results and found that CTBP1 is a top-ranked protein. It is molecular weight is 50kd. Therefore, we hypothesized that CTBP1 is the interacted protein with lin00492 in B cells. To verify this hypothesis, we preformed the Western blotting assays in pulled down proteins and the result indicated that CTBP1 was a direct binding partner of lnc00492 (Figure 5B). Consistently, the results of RNA immunoprecipitation (RIP) assays confirmed that anti-CTBP1 antibody can significantly enrich lnc00492 (Figure 5C). To determine which part of lnc00492 is involved in the interaction with CTBP1, we dissected lnc00492 into three regions (5′ middle and 3′) by PCR and detected their interaction with CTBP1 (Figure 5D). According to the results of RNA pull-down and Western blotting assays, we confirmed that the 1–625 nt motif sequence of lnc00492 is crucial for binding with CTBP1 (Figure 5E). These results suggest that lnc00492 can physically interact with CTBP1 in B cells.
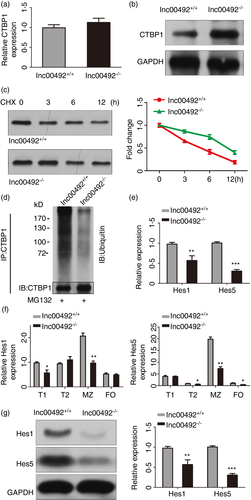
Lnc00492 regulates the ubiquitination of CTBP1 in B cells
To explore the mechanism of lnc00492 regulate the interact protein, we analysed the effects of lnc00492 on CTBP1 expression. The results indicated that knockout of lnc00492 do not affect CTBP1 mRNA levels (Figure 6A). We also found that knockout of CTBP1 does not affect lnc00492 mRNA levels (Figure S1). However, the protein level of CTBP1 was clearly increased in lnc00492 knockout B cells (Figure 6B). Hence, we hypothesized that lnc00492 interacts with CTBP1 and regulates its stability. Western blotting assays were employed to assess CTBP1 protein stability, and the results showed that CTBP1 protein levels were markedly up-regulated in lnc00492 knockout B cells treated with CHX (protein synthesis inhibitor) (Figure 6C). Moreover, because the degradation of CTBP1 protein in B cells was decelerated by lnc00492 knockout, we detected the level of ubiquitination of CTBP1 in lnc00492+/+ and lnc00492−/− mouse B cells. We found that compared with lnc00492+/+ group B cells, the ubiquitination level of CTBP1 was decreased in lnc00492−/− group B cells (Figure 6D). Taken together, our results show that the binding of lnc00492 to CTBP1 increased the ubiquitination level, leading to enhanced CTBP1 degradation. CTBP1 is a functional target protein of lnc00492 in B cells.
Lnc00492 deficiency blocks MZ B-cell development by regulating the Notch2 signalling pathway
Many research has indicated that the corepressor of the recombination signal binding protein CTBP1 can negatively regulate the Notch2 pathway. [25 The mainly function of CTBP1 is as an inhibitor in Notch2 signalling by targeting the transcription factor complex, resulting in the transcript inhibition of Hes family members. [26 The Notch2 signalling pathway is highly conserved across species and regulates cell proliferation, differentiation, fate and death. [27 Additionally, the Notch2 pathway plays an important role in B-cell development. Heterozygous knockout Notch2+/− mice demonstrated its function, leading to the decrease of MZ B cells. [28 Hence, we hypothesized that if CTBP1 is released in lnc00492−/− mouse MZ B cells, it may result in the expression inhibited of genes which are necessary in MZ B-cell development. To address this, we tested the effect of lnc00492 on the expression of Hes1/5 mRNAs. qRT-PCR results indicated that Hes1 and Hes5 significantly decreased in lnc00492−/− B cells (Figure 6E). With the same method, we also explored the expression of Hes1 and Hes5 in splenic B-cell subsets, especially in MZ B cells from lnc00492+/+ and lnc00492−/− mice. The results showed that there were a reductions in Hes1 and Hes5 (Figure 6F). Furthermore, we evaluated Hes1 and Hes5 expression in MZ B cells from lnc00492−/− and lnc00492+/+ mice by performing Western blotting assays. The results revealed that lnc00492 knockout significantly inhibited the protein levels of Hes1 and Hes5 (Figure 6G). Taken together, these results demonstrated that lnc00492 could decoy CTBP1 from the promoter binding sites of Hes family member genes and regulated the expression profile of Hes1/5.
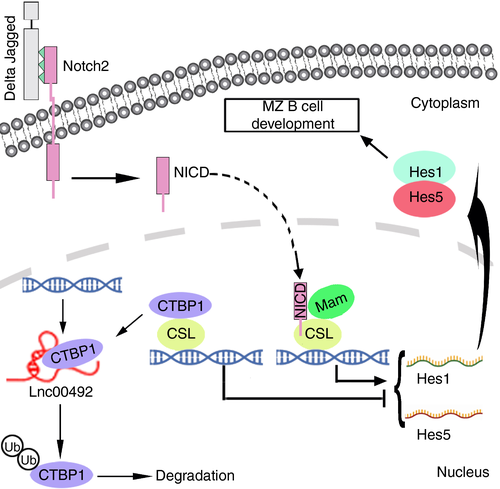
Based on our findings, we predict a model in which lnc00492 exerts its biological function by interacting with CTBP1 to activate its ubiquitin-mediated degradation and then participates in the transcription of Hes family member genes to increase the mRNA and protein levels of Hes1 and Hes5, thereby activating Notch2 signalling in MZ B cells (Figure 7).
DISCUSSION
Previous studies based on B-cell development focused on protein-coding genes. Many classical proteins have been shown to play an important role in B cells, which proves that the chromosomal region where they are located plays a key role in B-cell development. [29-31 However, the results of the Human Genome Project show that only 2% of RNA in the body is translated and folded into proteins. [32-34 These results all indicate that non-coding RNAs play an irreplaceable role in B-cell development. An increasing number of studies have revealed that miRNAs play a significant role in B-cell development. However, very little is known about the effect of lncRNAs on B-cell development. In this study, we identified a novel long non-coding RNA Gme00492, which is located on chr11 (start: 114204362; end: 114244555; - stand), and 7380 nt in length. We confirmed that lnc00492 is up-regulation in the B-cell development, which peaked at MZ B-cell subset. These results suggest that lnc00492 has an important role in B cells.
To determine whether the high expression of lnc00492 is related to the normal development of B cells, we constructed the lnc00492-deficient mouse model and examined the development of B cells in mice. Consistent with our hypothesis, the experimental results showed that compared with lnc00492+/+ mice, in the spleen the MZ B-cell subset of lnc00492−/− mice were significantly reduced, while the T1 and T2 subsets were significantly blocked. However, there were no different in lymphoid and peripheral blood MZ B cells. Furthermore, the proliferation and apoptosis of MZ B-cell also no changed. Hence, those results suggest that the decrease of MZ B cell may be caused by developmental defects in the spleen. Surprisingly, the development of B cells was largely unaffected in bone marrow.
Interestingly, we detected that lnc00492 localized in the nucleus. According to existing research, the nuclear location of lncRNA modulates chromatin organization, RNA processing and transcription. [35-37 LncRNAs can negatively or positively affect the expression of target genes. For example, lncRNA PDPK2P can as a factor interact with proteins. In HCC cells, mechanism studies have shown that it regulates HCC cell proliferation and apoptosis by promoting the transcription of PDK1· [38 The novel lncRNA RP11-279C4·1 plays an important role in glioma cells. Further studies have shown that it regulates the cycle and apoptosis of glioma cells by inducing the differentiation of CBX3· [39 Hence, we hypothesize that lnc00492 may interact with transcription factors as a decoy RNA in the B-cell nucleus. RNA pull-down assays and RIP assays confirmed this hypothesis. We demonstrated the regulate mechanism by which nuclear lnc00492 inhibited MZ B-cell development by directly interacting with CTBP1.
CTBP1 is a key gene in the Notch2 signalling pathway, and a well-known transcriptional corepressor factor, locating in the nucleus of cells. [40 CTBP1 plays an important role in B cells by assembling polycomb complexes to silence target gene expression through hypermethylation. [41 Our results showed that deletion of lnc00492 resulted in significant increases in CTBP1 protein levels. Interestingly, the transcript level of CTBP1 was similar. Furthermore, we demonstrated that lnc00492-deficient MZ B cells showed a reduction in Hes1 and Hes5 expression, which were the target genes of Notch2. Taken together, these data indicate that lnc00492 posttranslationally regulated CTBP1 expression by affecting its ubiquitin-protein media degradation pathway. Lnc00492 promote the development of MZ B cells by activating Notch2 signalling.
In summary, we successfully constructed a lnc00492−/− mouse model and proved that the deletion of lnc00492 seriously affects the development of MZ B cells. Lnc00492 is up-regulated in B-cell development and posttranslationally regulates the expression of CTBP1 by affecting its ubiquitin-protein media degradation pathway. High protein stability of CTBP1 decreased Hes1 and Hes5 mRNA levels, leading to inactivation of the Notch2 signalling pathway. These findings provide a novel regulatory axis between lnc00492-controlled Hes1 and Hes5 expression in B cells, which explains why lnc00492 is required for B-cell development.
ACKNOWLEDGEMENTS
This research was supported by the Graduate Research and Innovation Projects of Jiangsu Province (KYCX19_0114); special thanks to Ms Lu Chen (Center of Modern Analysis, Nanjing University, Nanjing, China) for their support with the mass spectrometry analysis.
CONFLICT OF INTEREST
No potential conflict of interest was reported by the authors.
AUTHOR CONTRIBUTIONS
L.Y., D.W. and F.W. conceived and designed the experiments and wrote the paper. F.W. and D.C. performed the experiments, analysed the data and wrote the paper. D.C. generated the lnc00492-/- and lnc00492+/+ mice. B-cell development analyses were performed by F.W., D.Z., Y.S. and Y.L. Flow cytometry measurements were performed by B.Z., and L.C. F.W. and L.Y. discussed the data.




