Assessment of species limits in African ‘brown buntings’ (Emberiza, Passeriformes) based on mitochondrial and nuclear sequence data
Abstract
We estimated a phylogeny for 10 taxa currently placed in four polytypic species that collectively encompass the African ‘brown buntings’: Cape Bunting Emberiza capensis, Cinnamon-breasted Bunting Emberiza tahapisi, Lark-like Bunting Emberiza impetuani and House Bunting Emberiza striolata. We made use of the mitochondrial cytochrome b gene and the nuclear introns 6–7 of ornithine decarboxylase (ODC), and intron 2 of myoglobin. There was substantial cytochrome b sequence divergence between taxa currently treated as conspecific: sahari vs. striolata (2.6–3.1% (uncorrected-p); 3.0–3.6% (HKY + I)), and goslingi vs. tahapisi (4.4–4.7% (uncorrected-p); 5.4–5.9% (HKY + I)). The degree of divergence of the nuclear loci among taxa was limited, and these loci lacked reciprocal monophyly, most likely as a consequence of incomplete lineage sorting. A single representative of the taxon septemstriata, generally treated as a member of the dark-throated tahapisi group, here appears to be genetically consistent with the grey-throated goslingi, and may be of hybrid origin. All other taxa allocated to E. striolata and E. tahapisi make up four reciprocally monophyletic groups consistent with sahari, striolata, tahapisi and goslingi, respectively. The extent of genetic evidence suggests that these taxa have been evolving as separate evolutionary lineages for a long time. This is further manifested in several morphological and vocal characteristics described previously, and we propose that these divergent taxa be treated as separate species: Cinnamon-breasted Bunting Emberiza tahapisi, Gosling's Bunting Emberiza goslingi, Striolated Bunting Emberiza striolata and House Bunting Emberiza sahari. We do not propose any taxonomic changes regarding Emberiza impetuani or Emberiza capensis.
The African ‘brown buntings’, Cape Bunting Emberiza capensis, Cinnamon-breasted Bunting Emberiza tahapisi, Lark-like Bunting Emberiza impetuani and House Bunting Emberiza striolata, are medium-sized seed-eating passerines that are distributed over much of Africa, with the last-named species occurring outside the continent as far east as India. These taxa generally prefer arid regions or hilly country with sparse vegetation, although Cinnamon-breasted Bunting may occur in open forest. Most areas are inhabited by a single species, but regions of geographically overlapping ranges are found in southern Africa and in the Sahel of northern Africa. All taxa in the complex lack white in the tail, unlike most other buntings, and all taxa with the exception of Lark-like Bunting and the ‘NW African group’ of House Bunting Emberiza striolata sahari have prominent pale and dark stripes on the crown and ear-coverts (Byers et al. 1995, Fry & Keith 2004).
Hall and Moreau (1970) suggested that Cape, Cinnamon-breasted, Lark-like and House Bunting form a species complex together with Rock Bunting Emberiza cia. Alström et al. (2008) confirmed that Cape, Cinnamon-breasted, Lark-like and House Bunting form a monophyletic group, but found that Rock Bunting is not closely related to this clade. The African ‘brown buntings’, together with Socotra Bunting Emberiza socotrana, have been placed in the genus Fringillaria, erected by Swainson 1837, although most authors treat these taxa as part of the genus Emberiza (e.g. Paynter & Storer 1970, Sibley & Monroe 1990, Byers et al. 1995, Dickinson 2003, Fry & Keith 2004). The molecular phylogeny of Alström et al. (2008) found that the African ‘brown bunting’ clade, together with the African ‘yellow buntings’, may be sister to the core Emberiza clade. Thus, the option of using the name Fringillaria is available for the African buntings. The Socotra Bunting, which is endemic to Socotra Island, shares many morphological similarities with the African ‘brown buntings’ and may be part of this clade. However, it has never been included in a molecular phylogeny.
The House Bunting occurs patchily in arid habitats from Mali and Morocco to westernmost India. In Africa, it occurs south to the Sahel, extending in the east as far south as Lake Turkana in Kenya. Several subspecies have been described, although there is considerable disagreement among authors regarding their validity (Vaurie 1956, Byers et al. 1995, Kirwan & Shirihai 2007). In particular, there is uncertainty surrounding the taxonomic status of the subsaharan populations, which are poorly studied and patchily distributed. Some of these taxa have been regarded as intergrades between the otherwise allopatric subspecies striolata and sahari, which has been taken as evidence for conspecificity between these two taxa. Kirwan and Shirihai (2007) considered most subsaharan populations to be referable to either the subspecies striolata or sahari, and only considered three groups to be diagnosable (striolata, sahari and saturiator/jebelmarrae). Disregarding the last of these for lack of information, Byers et al. (1995), Kirwan and Shirihai (2007) and Mullarney et al. (2010) described differences in vocalizations and ecology between the subspecies striolata and sahari, and Kirwan and Shirihai (2007) proposed that they be treated as separate species, which has been adopted by the IOC Checklist (Gill & Donsker 2011).
Cinnamon-breasted Bunting occurs across large parts of Africa south of the Sahara and marginally extends into the southern Arabian Peninsula. This species is only absent from the equatorial forest region, at high altitudes on mountains, and in deserts (Byers et al. 1995, Fry & Keith 2004). Five subspecies are recognized by Byers et al. (1995). The male of the most widely distributed subspecies tahapisi is strikingly striped black and white on the head, with uniformly black throat and upper breast. This is also the case in the three subspecies septemstriata, insularis and arabica. However, the subspecies goslingi, which is distributed from Senegal to western Sudan, differs from the others in having almost entirely rufous remiges and in the adult male a uniformly pale grey throat (e.g. Byers et al. 1995, Fry & Keith 2004). The differences in vocalizations and ecology between goslingi and other taxa are little known, but Osiejuk (2011) found that certain syllables characteristic of the song of a population in northwestern Cameroon (goslingi) was also present in a recording from Nigeria (goslingi), but not in available recordings from Ethiopia, Yemen, Zimbabwe or Lesotho (tahapisi group). Although the scope of Osiejuk (2011) was not a taxonomic study and no detailed comparison was made, the results nevertheless indicate that there may exist consistent differences in song between the goslingi and the tahapisi groups.
Lark-like Bunting occupies the southernmost parts of Africa, where it inhabits stony areas with sparse vegetation cover. It differs from the other species in this group by lacking the dark stripes on the sides of the head. Three poorly differentiated subspecies have been described. Cape Bunting is confined to southern Africa, where it inhabits rocky areas with sparse vegetation cover. It is geographically variable, ranging from very pale to medium brown, with two outlying geographically isolated forms, which are characterized by their very dark plumage.
We present a phylogenetic analysis of the African ‘brown bunting’ complex, with special focus on species limits in the Cinnamon-breasted and House Bunting species complexes using Bayesian inference, based on a dataset comprising one mitochondrial (cytochrome b) and two nuclear loci (ODC introns 6–7 and myoglobin intron 2).
Methods
Tissue samples (Table 1) were collected from the following taxa: Emberiza striolata sahari (Tunisia, n = 1), Emberiza striolata sahari/sanghae (Mauretania, n = 4), Emberiza striolata striolata (Israel, n = 2), Emberiza tahapisi tahapisi (Malawi, n = 2), Emberiza tahapisi tahapisi (South Africa, n = 1), Emberiza tahapisi arabica (Oman, n = 1), Emberiza tahapisi septemstriata (Eritrea, n = 1), Emberiza tahapisi goslingi (Nigeria, n = 3), Emberiza impetuani sloggeti (South Africa, n = 1), Emberiza capensis capensis (South Africa, n = 1), Emberiza capensis media (South Africa, n = 1) and Emberiza capensis smithersi (Zimbabwe, n = 1). Golden-breasted Bunting Emberiza flaviventris and Yellowhammer Emberiza citrinella were used as outgroups, based on the results of Alström et al. (2008). Samples of Grassland Sparrow Ammodramus humeralis, Rock Bunting E. cia and Cabanis's Bunting Emberiza cabanisi were added a posteriori to investigate a deviating sequence in the myoglobin dataset. DNA was obtained from either blood samples or feathers collected from live birds, except for E. capensis smithersi and E. tahapisi septemstriata, for which toe-pads from museum specimens were used. The blood samples were extracted using a Qiagen Blood Kit (Qiagen, Hilden, Germany), according to the manufacturer's recommendations. Feathers and toe-pads were extracted either with a Qiaamp Mini Kit or Qiaamp DNEasy Kit, following the manufacturer's recommendations, with the exception that 30 μL 0.1% dithiothreitol was added to the first incubation step to dissolve the feathers and toe-pads and thereby increase the DNA yield.
| Taxon | Locality | Museum no. | Regions | GenBank no. | Documentation |
|---|---|---|---|---|---|
| Ammodramus humeralis xanthornus | Paraguay | NRM 976701 | myo | JX515370 | Complete skeleton, photo |
| Emberiza cabanisi cabanisi | Cameroon | VH, uncatalogued DZUG U773 | myo | JX515372 | – |
| Emberiza capensis capensis | Cape prov., South Africa | PFI, uncatalogued DZUG U1062 | cyt b ODC myo | EU325765 EU325823 JX515375 | – |
| Emberiza capensis media | Orange Free State, South Africa | NMB GA85844 | cyt b ODC myo | JX515340 JX515355 JX515376 | – |
| Emberiza capensis smithersi | Zimbabwe | AMNH SKIN 788876 | cyt b ODC myo | JX515354 JX515369 JX515393 | Specimen voucher |
| Emberiza cia cia | Spain (b) | NRM 20076340 | myo | JX515371 | Wing |
| Emberiza citrinella citrinella | Sweden | NRM 20076343 | cyt b ODC myo | EU325753 EU325811 JX515373 | – |
| Emberiza flaviventris ssp. | Captive | UMMZ 233274 | cyt b ODC myo | EU325766 EU325824 JX515374 | Wing, skeleton |
| Emberiza goslingi | Nigeria | DZUG U795 | cyt b ODC myo | JX515343 JX515358 JX515380 | – |
| Emberiza goslingi | Nigeria | DZUG U1067 | cyt b ODC myo | JX515341 JX515356 JX515378 | – |
| Emberiza goslingi | Nigeria | DZUG U1068 | cyt b ODC myo | JX515342 JX515357 JX515379 | – |
| Emberiza impetuani sloggetti | Orange Free State, South Africa | NMB GA85845 | cyt b ODC myo | EU325764 EU325822 JX515377 | – |
| Emberiza sahari sahari | Mauretania | DZUG U1069 | cyt b ODC myo | JX515344 JX515359 JX515381 | Photo |
| Emberiza sahari sahari | Mauretania | DZUG U1070 | cyt b ODC myo | JX515345 JX515360 JX515382 | Photo |
| Emberiza sahari sahari | Mauretania | DZUG U1071 | cyt b ODC myo | JX515346 JX515361 JX515383 | Photo |
| Emberiza sahari sahari | Mauretania | DZUG U1072 | cyt b ODC myo | JX515347 JX515362 JX515384 | Photo |
| Emberiza sahari sahari | Tunisia | DZUG U1064 | cyt b ODC myo | JX515348 JX515363 JX515385 | – |
| Emberiza striolata striolata | Israel | NRM 20076357 | cyt b ODC myo | EU325762 EU325820 JX515386 | Photo |
| Emberiza striolata striolata | Israel | DZUG U1065 | cyt b ODC myo | JX515349 JX515364 JX515387 | Photo |
| Emberiza tahapisi septemstriata | Eritrea | ZMUC 71.190 | cyt b ODC myo | JX515353 JX515368 JX515392 | Specimen voucher |
| Emberiza tahapisi tahapisi | Malawi | NRM 20076359 | cyt b ODC myo | EU325763 EU325821 JX515389 | Photo |
| Emberiza tahapisi tahapisi | Malawi | DZUG U1345 | cyt b ODC myo | JX515351 JX515366 JX515390 | Photo |
| Emberiza tahapisi tahapisi | South Africa | DZUG U521 | cyt b ODC myo | JX515350 JX515365 JX515388 | – |
| Emberiza tahapisi arabica | Oman | DZUG U792 | cyt b ODC myo | JX515352 JX515367 JX515391 | – |
We sequenced three loci: the mitochondrial cytochrome b gene (cyt b), introns 6 to 7 of the nuclear ornithine decarboxylase gene (ODC) and intron 2 of the nuclear myoglobin gene (myo). PCR-amplification and sequencing of the cytochrome b gene followed the protocols described in Olsson et al. (2005); introns 6–7 of the ODC gene followed Friesen et al. (1999), Allen and Omland (2003) and Irestedt et al. (2006); and for intron 2 of myoglobin we followed Olsson et al. (2005). Except for the toe-pad samples, cyt b was amplified as one fragment to decrease the risk of PCR-amplifying nuclear pseudogenes (Zhang & Hewitt 1996, Sorensen & Quinn 1998). The toe-pads were PCR-amplified in short fragments using a variety of specifically designed primer pairs: details are available from the authors. Sequences were aligned using megalign 4.03 in the DNASTAR package (DNAstar Inc. Madison, WI, USA); some manual adjustment was necessary for the ODC sequences.
Phylogenies were estimated by Bayesian inference (BI) using MrBayes 3.2 (Huelsenbeck & Ronquist 2001, 2005, Ronquist & Huelsenbeck 2003). For BI, all loci were analysed both separately (single-locus analyses, SLAs) and concatenated (BI of the concatenated dataset). The myo dataset was analysed twice to investigate the nature of a deviating sahari sequence. In the second BI, three outgroups were added. In the BI of the concatenated dataset, the data were partitioned such that the non-coding nuclear introns and the protein-coding cyt b were analysed separately, using rate multipliers to allow different rates for the different partitions (Ronquist & Huelsenbeck 2003, Nylander et al. 2004). For BI, two simultaneous runs, each with four Metropolis-coupled MCMC chains with incremental heating temperature 0.1, were run for 5x107 generations and sampled every 1000 generations. The first 20% of the generations were discarded after manual inspection for stationarity of chain likelihood values and the average standard deviation of split frequencies in MrBayes. The posterior probability was estimated from the remaining 4x107 generations.
The choice of model for the partitions in the BI of the concatenated dataset was determined based on the Bayesian information criterion (Schwarz 1978) calculated in jModeltest (Posada 2008). For all loci, posterior probabilities were calculated under the (HKY) model (Hasegawa et al. 1985), and for the cyt b and myo data also assuming rate variation across sites according to a discrete gamma distribution with four rate categories (Γ; Yang 1994).
Pairwise genetic distances for cyt b (excluding outgroups) were calculated in paup* under the distance criterion (Swofford 2002) using both uncorrected-p and maximum likelihood distances, following the recommendations of Fregin et al. (2012) to use ‘complete deletion’ homologous sequences (same length) and optimal substitution models. The optimal substitution model for the modified cyt b dataset corresponded to the HKY + I model (Hasegawa et al. 1985, Gu et al. 1995). The proportion of invariable sites was obtained from a BI of the modified cyt b dataset, since paup* is not able to estimate these parameters under the distance criterion.
Results
Including the outgroup taxa, the aligned cyt b dataset comprised 1076 characters, of which 157 (14.6%) were parsimony informative; ODC comprised 705 characters, 13 (1.8%) informative; and myo 716 characters, 27 (3.8%) parsimony informative. The total dataset comprised 2497 characters. No unexpected start or stop codons that could indicate the presence of nuclear copies are present in the cyt b sequences.
The tree based on the concatenated sequences (Fig. 1a) is well supported and entirely congruent with the SLA of the cyt b dataset (Fig. 1b). In the tahapisi clade, the samples of tahapisi from Malawi and South Africa form a clade which is sister to the sample of arabica from Oman. These are sisters to a clade including the samples of goslingi from Nigeria and septemstriata from Eritrea. The striolata and sahari samples are divided into two well-supported clades, in line with current taxonomy.
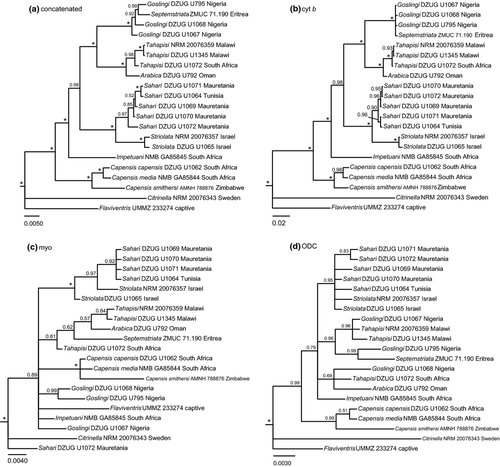
The SLAs of the mitochondrial and nuclear datasets show relatively few conflicts receiving posterior probabilities (PP) ≥ 0.95 (Fig. 1b–d). In the cyt b SLA (Fig. 1b), all clades are in agreement with the current taxonomy (Dickinson 2003), except for the position of septemstriata. The myo SLA (Fig. 1c) exhibits one instance of direct conflict, concerning one sample of sahari (DZUG 1072), which in the myo SLA is placed outside the clade including all other sahari and striolata samples, with PP 1.00. In the myo SLA, it is placed outside the entire clade of African ‘brown buntings’, although support is insufficient (PP 0.89). In the second BI of the myo dataset (Supporting Information Fig. S1), with distant outgroups added, the sahari (DZUG 1072) sequence is part of the sahari/striolata clade with PP 0.98.
In the ODC SLA (Fig. 1d), the topology of the ingroup receives mixed support. The striolata/sahari clade is well supported but unresolved internally. The taxa tahapisi, goslingi and septemstriata are part of one well-supported clade, to the exclusion of an insufficiently supported clade that includes tahapisi, goslingi and arabica. The three samples in the capensis group form a well-supported clade.
Pairwise genetic divergences are provided in Table 2 and Table S1. The estimated proportion of invariable sites was 0.7506 in the modified cyt b dataset.
| capensis capensis | capensismedia | capensis smithersi | impetuani | goslingi DZUG U1067 | goslingi Ho0205 DZUG U1068 | goslingi Ha0205 DZUG U795 | septemstriata ZMUC 71.190 | sahari MauretaniaDZUG U1069 | sahari MauretaniaDZUG U1070 | sahari MauretaniaDZUG U1071 | sahari MauretaniaDZUG U1072 | sahari Tunisia DZUG U1064 | striolata Eilat03 NRM 20076357 | striolata Eilat04 DZUG U1065 | tahapisi tahapisi SA DZUG U521 | tahapisi tahapisi Malawi1 NRM 20076359 | tahapisi tahapisi Malawi2 DZUG U1345 | tahapisi arabica Oman DZUG U792 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| capensis capensis | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| capensis media | 0.9 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| capensis smithersi | 2.6 | 2.6 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| impetuani | 9.5 | 10.0 | 9.2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| goslingi DZUG U1067 | 9.5 | 9.9 | 9.9 | 9.2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| goslingi DZUG U1068 | 9.7 | 10.2 | 10.1 | 9.4 | 0.1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| goslingi DZUG U795 | 9.7 | 10.2 | 10.1 | 9.4 | 0.1 | 0.2 | – | – | – | – | – | – | – | – | – | – | – | – | – |
| septemstriata ZMUC 71.190 | 9.7 | 10.2 | 10.1 | 9.4 | 0.1 | 0.2 | 0.2 | – | – | – | – | – | – | – | – | – | – | – | – |
| sahari DZUG U1069 | 7.9 | 8.3 | 8.1 | 8.7 | 7.1 | 7.3 | 7.3 | 8.1 | – | – | – | – | – | – | – | – | – | – | – |
| sahari DZUG U1070 | 8.1 | 8.6 | 8.3 | 8.9 | 6.9 | 7.1 | 7.1 | 7.1 | 0.1 | – | – | – | – | – | – | – | – | – | – |
| sahari DZUG U1071 | 7.7 | 8.1 | 7.9 | 8.5 | 6.9 | 7.1 | 7.1 | 7.1 | 0.3 | 0.5 | – | – | – | – | – | – | – | – | – |
| sahari DZUG U1072 | 8.1 | 8.6 | 8.3 | 8.9 | 6.9 | 7.1 | 7.1 | 7.1 | 0.1 | 0.0 | 0.5 | – | – | – | – | – | – | – | – |
| sahari DZUG U1064 | 7.5 | 7.9 | 7.7 | 8.7 | 6.7 | 6.9 | 6.9 | 6.9 | 0.5 | 0.6 | 0.1 | 0.6 | – | – | – | – | – | – | – |
| striolata NRM 20076357 | 10.0 | 10.5 | 9.7 | 9.9 | 9.0 | 9.2 | 9.2 | 9.2 | 3.2 | 3.3 | 3.0 | 3.3 | 3.2 | – | – | – | – | – | – |
| striolata DZUG U1065 | 10.4 | 10.9 | 10.1 | 10.3 | 9.4 | 9.6 | 9.6 | 9.6 | 3.4 | 3.6 | 3.3 | 3.6 | 3.4 | 0.2 | – | – | – | – | – |
| tahapisi DZUG U521 | 8.9 | 8.9 | 8.4 | 8.6 | 5.7 | 5.9 | 5.9 | 5.9 | 7.6 | 7.4 | 7.4 | 7.4 | 7.2 | 9.1 | 9.6 | – | – | – | – |
| tahapisi NRM 20076359 | 8.9 | 8.9 | 8.4 | 8.6 | 5.7 | 5.9 | 5.9 | 5.9 | 7.6 | 7.4 | 7.4 | 7.4 | 7.2 | 9.1 | 9.6 | 0.0 | – | – | – |
| tahapisi DZUG U1345 | 8.9 | 8.9 | 8.4 | 8.6 | 5.7 | 5.9 | 5.9 | 5.9 | 7.6 | 7.4 | 7.4 | 7.4 | 7.2 | 9.1 | 9.6 | 0.0 | 0.0 | – | – |
| arabica DZUG U792 | 9.0 | 9.0 | 9.0 | 8.7 | 5.4 | 5.6 | 5.6 | 5.6 | 7.3 | 7.1 | 7.1 | 7.1 | 6.9 | 9.2 | 9.6 | 1.2 | 1.2 | 1.2 | – |
Discussion
Our data provide information about the phylogenetic patterns in the African ‘brown bunting’ complex, but the interpretation is not unproblematic. The trees resulting from the concatenated dataset and the SLA of the mitochondrial cyt b (Fig. 1a,b) are congruent with current taxonomy in that most taxa are monophyletic. The exception to this is the placement of septemstriata in the goslingi clade. Byers et al. (1995) suggested that the former may be of hybrid origin, based on variable amounts of intermediate traits between goslingi and tahapisi, which is not rejected by the present analyses. Furthermore, the topologies of the two nuclear introns complicate the picture. Nuclear introns become fixed at a slower rate than mitochondrial markers due to their larger effective population size, which increases the likelihood of incomplete lineage sorting in the case of recent divergence. Nuclear introns would thus be expected to show reciprocal monophyly later than mitochondrial markers would. Moreover, in the ODC dataset, only one single nucleotide polymorphism (SNP) within the sahari/striolata clade is parsimony informative, differing only between sahari samples. In the same dataset, only five SNPs are parsimony informative within the tahapisi/goslingi clade, none of which segregates between the taxa. Accordingly, the nuclear data contain too little phylogenetic information to be able to resolve the relationships.
For the individual of sahari (DZUG 1072) that is placed outside the entire African clade in the myo SLA (Fig. 1c), 11 SNPs (1.6%) are parsimony informative compared with Yellowhammer, with which it shares a unique 8-bp indel, whereas only three SNPs (0.4%) are parsimony informative within the sahari/striolata clade. However, the amount of divergence in this sample is so large that introgression from a species not included in the dataset could be suspected. Myo sequences of other buntings of the genus Emberiza were not available, but we sequenced Cabanis's Bunting from the African ‘yellow bunting’ group and Rock Bunting, which were suggested to be part of the same group as the African ‘brown buntings’ by Hall and Moreau (1970), to investigate the origin of this rogue sequence. Inclusion of these species, and an outgroup taxon from outside the Emberiza clade, resulted in a phylogeny where the DZUG 1072 myo sequence is sister to the sahari/striolata clade (Fig. S1), with PP 0.98, rejecting the hypothesis of introgression.
Taxonomic implications
The uncorrected cyt b divergence between striolata and sahari varies between 3.0 and 3.6%, whereas the divergence between tahapisi and goslingi varies between 5.4 and 5.9% (Table 2). These values are somewhat smaller than the pairwise comparisons between the generally recognized species in this clade: the minimum is 6.9% between sahari and arabica, and maximum 10.3% between impetuani and striolata, and 9.6% between tahapisi and striolata (Table 2). However, in many cases, taxa showing similar levels of divergence as between striolata and sahari, and tahapisi and goslingi, respectively, have been ranked as separate species (e.g. Martens et al. 2004, Li et al. 2006, Qu et al. 2006, Brambilla et al. 2008, Bowie et al. 2009). Although we do not consider the amount of divergence as such a useful measure of species status, it does indicate that the taxa in each of these two pairs have been evolving as separate lineages for a substantial period of time.
The lack of reciprocal monophyly in the nuclear intron sequences of the taxa in question may be viewed as a problem. However, the conflicting topologies are not strongly supported and are probably best regarded as an effect of incomplete lineage sorting in the ongoing process of recent lineage divergence.
The concordant differences in plumage, vocalizations and ecology between striolata and sahari (Byers et al. 1995, Kirwan & Shirihai 2007, Mullarney et al. 2010) are independent evidence that gene flow has been absent or reduced for a considerable period of time, and we agree with Kirwan and Shirihai (2007) that striolata and sahari should be regarded as different species. It should be noted that the taxonomic validity of the populations of the striolata complex in Mali, and taxonomic status and possible interactions of populations in eastern Chad and Sudan, still remain undetermined. It has been suggested that the appearance of at least some of these populations, which are in some respects morphologically intermediate between striolata and sahari, may be due to hybrid origin (Byers et al. 1995).
Within the E. tahapisi complex there is a considerable divergence in morphology (Byers et al. 1995) between the representatives of the dark-throated forms (here represented by tahapisi, septemstriata and arabica), and the grey-throated goslingi. The divergence in cyt b between tahapisi and goslingi is higher than between striolata and sahari, and although differences in ecology and vocalizations remain insufficiently known, the congruent divergence in plumage and DNA is strong evidence that they represent separate evolutionary lineages. The single sample of the dark-throated arabica differs by about 1.2% (HKY + I) from tahapisi, indicating that it represents an evolutionarily separate lineage, but further study is needed to understand better the diversification within the dark-throated tahapisi group, which also includes insularis and septemstriata. The latter taxon is morphologically variable, exhibiting traits intermediate between tahapisi and goslingi, and was suggested to be of hybrid origin by Byers et al. (1995). The head pattern is usually similar to tahapisi, whereas the amount of rufous in the remiges is more typical of goslingi. Our single sample is morphologically most similar to tahapisi, but is placed in the goslingi clade with high support both in the phylogenies based on the concatenated dataset and for cyt b. The genetic distance (HKY + I) between the septemstriata sequence and the three goslingi samples is 0.1–0.2%, whereas it is 5.6–5.9% compared with tahapisi and arabica. The nuclear data are inconclusive. Our single individual does not resolve the question, but the hypothesis that septemstriata represents a hybrid population is not contradicted by our data.
If septemstriata indeed represents a hybrid swarm between goslingi and tahapisi, interpretation of their species status would be affected according to species definitions requiring intrinsic reproductive isolation sensu Mayr (1963). However, we do not regard localized hybridization to be of paramount importance in the determination of species limits. There are many examples of species that hybridize locally, and birds appear to be slow in evolving post-zygotic reproductive barriers (Grant & Grant 1992, Price & Bouvier 2002, Price 2008). Instead, we consider the morphological and phylogenetic evidence shows that the main population centres of the goslingi and the tahapisi groups have evolved as separate evolutionary units for a substantial period of time, and that the level of their overall divergence suggests that they will remain distinct, which is more important in this respect (Helbig et al. 2002). Furthermore, the ability of two separate taxa to interbreed is in most cases probably a hold-over from the time when they were part of a single ancestral population, rather than a capacity that has evolved de novo. Using such a plesiomorphic condition to group two taxa together would not be consistent with cladistic methodology.
The genetic divergence between the two southern Cape Bunting taxa is slight (0.9%, HKY + I) (Table 2), but the morphologically distinct smithersi differs by 2.6% (HKY + I) from each of these, suggesting substantial time in isolation. However, there are many subspecies of the Cape Bunting remaining to be studied by molecular methods, and we do not recommend at present any taxonomic changes based on these results.
The combined evidence of the phylogenetic analyses presented here, and the concordant differences in plumage, vocalizations and ecology (Byers et al. 1995, Fry & Keith 2004, Kirwan & Shirihai 2007, Mullarney et al. 2010, Osiejuk 2011), indicate long-standing divergence within both House Bunting and Cinnamon-breasted Bunting, and we propose the recognition of four separate species: Cinnamon-breasted Bunting Emberiza tahapisi, Gosling's Bunting Emberiza goslingi, Striolated Bunting Emberiza striolata and House Bunting Emberiza sahari (Table 3). However, our sample size is limited and there is shortage of ecological information, particularly concerning goslingi, and further studies will provide deeper insight. In particular, the taxonomic status of the subsaharan populations in the E. striolata complex are in need of further study, as is the possible hybrid origin of septemstriata.
|
Emberiza striolata striolata Lichtenstein 1823, from India to coastal Sudan and Eritrea, and north to Israel. We provisionally include the taxa saturiator and jebelmarrae in this species, pending further studies, but refrain from suggesting any taxonomic action at this point: Emberiza striolata saturiator Sharpe 1901, southern Sudan, Ethiopia and south to Kenya. Emberiza striolata jebelmarrae Lynes 1920, western Sudan. Emberiza sahari sahari Levaillant 1850, from Morocco across Africa to northwest Libya. We provisionally include the taxa theresae and sanghae in this species, pending further studies, although the validity of both have been questioned (Kirwan & Shirihai 2007, and references therein): Emberiza sahari theresae Meinertzhagen 1939, described from southwest Morocco (based on only four individuals). Emberiza sahari sanghae Traylor 1960, described from southern Mali, based on only two individuals. Perhaps this taxon is also in northern Senegal and Mauretania, but the results of this study do not support the distinction of the Mauretanian population from sahari in Tunisia. We agree with Kirwan and Shirihai (2007) that the Mauretanian population should be considered a junior synonym of sahari, but take no position regarding the status of the population from southern Mali. Emberiza tahapisi tahapisi A. Smith 1836, most of Africa, from southern Cameroon and Ethiopia southwards. Emberiza tahapisi arabica Lorenz & Hellmayr 1902, southern Arabian peninsula. Emberiza tahapisi insularis Ogilvie-Grant & Forbes 1899, Socotra Island. We provisionally include the taxon septemstriata in this species, pending further studies, although Byers et al. proposed that it may represent a hybrid population between tahapisi and goslingi and our own data do not reject this suggestion. Emberiza tahapisi septemstriata Rüppel 1840, Sudan east of the Nile, northern Ethiopia. Emberiza goslingi monotypic Alexander 1906, Gambia to Sudan west of the Nile. |
Samples were generously provided by Penn Lloyd (Emberiza capensis capensis from South Africa), Dawie de Swardt (Emberiza capensis media and Emberiza impetuani from South Africa), Bob Medland (tahapisi from Malawi), David Mindell (Emberiza flaviventris), Ross McGregor (goslingi from Nigeria), Heiko Schmaljohann (sahari from Mauretania), Claire Spottiswoode (tahapisi from South Africa) and Lars Svensson (arabica from Oman). We are grateful to Jon Fjeldså and Jan Bolding Kristensen at the Zoological Museum of the University of Copenhagen, and Tom Trombone and Paul Sweet at the American Museum of Natural History for generously granting tissue loans from the collections of their respective museums. We are grateful for the constructive criticism on an earlier version of this manuscript by two anonymous reviewers and Rauri Bowie. The Swedish Research Council provided financial support (grant no. 621-2006-3194 to U.O.), and we gratefully acknowledge the Chinese Academy of Sciences Visiting Professorship for Senior International Scientists (2011T2S04) to P.A.




