A reappraisal of the systematic affinities of Socotran, Arabian and East African scops owls (Otus, Strigidae) using a combination of molecular, biometric and acoustic data
Abstract
We investigated phylogenetic relationships among Otus scops owls from Socotra Island, the Arabian Peninsula and East Africa using molecular, vocalization and biometric data. The Socotra Scops Owl Otus senegalensis socotranus, currently treated as a subspecies of the African Scops Owl Otus senegalensis, is more closely related to the Oriental Scops Owl Otus sunia and to the endemic Seychelles Scops Owl Otus insularis. Considerable mitochondrial genetic distance and significant morphological differentiation from its two closest relatives, as well as its distinctive vocalizations compared with O. insularis, strongly support recognition of Socotra Scops Owl as a full species. Unexpectedly, two taxa from the Arabian Peninsula, Pallid Scops Owl Otus brucei and African Scops Owl Otus senegalensis pamelae, represent very distinct lineages; O. brucei is basal to a clade that includes taxa found in the Indo-Malayan region and on Indian Ocean islands. In contrast, O. s. pamelae occupies a well-supported basal position within a clade of continental Afro-Palaearctic taxa. The uncorrected-p genetic distance between O. s. pamelae and its closest relatives (other populations of senegalensis from mainland Africa) is c. 4%. As O. s. pamelae is also well differentiated phylogenetically, morphologically and vocally from O. s. senegalensis, we recommend its elevation to species status, as Otus pamelae. Among mainland African O. senegalensis subspecies, Ethiopian populations appear to represent the most divergent lineage, whereas other lineages from Somalia, Kenya and South Africa are poorly differentiated. The large genetic distance between the Ethiopian haplotype and other African haplotypes (3.2%) suggests that the Ethiopian Otus may represent a cryptic taxon, and we recommend that more individuals be sampled to assess the taxonomic status of this population.
Scops owls (Otus, Strigidae) represent a genus of small to medium-sized nocturnal birds with strong colonizing capability, suggested by their presence on small and remote islands. Despite high species richness, the morphological diversity encountered within the genus is limited, with the occurrence of only two primary plumage colour types: rufous or grey. This lack of major plumage differences is common among members of the Strigidae and is usually explained by the need to remain inconspicuous during the day as a defence against predators or from being mobbed by other birds (Marks et al. 1999). As a consequence of their highly conserved morphology, it is unsurprising that the systematics of the genus Otus, as traditionally defined, have been challenged by molecular data (Fuchs et al. 2008, Wink et al. 2009, Miranda et al. 2011). These studies suggested not only that the genus Otus is polyphyletic, because the White-faced Scops Owl Otus leucotis is sister to the genus Asio, whereas the New World Screech Owls have affinities with the genera Bubo, Strix and the Asio–Otus leucotis clade (Fuchs et al. 2008), but also that species limits in some groups must be reconsidered due to either divergent mtDNA sequences (Miranda et al. 2011) or paraphyly of traditionally recognized species (e.g. Collared Scops Owl O. lettia; Fuchs et al. 2008).
As currently understood, the genus Otus has its centre of diversity in the Palaearctic and Oriental (Indo-Malayan) regions (26 species). Secondary radiations have occurred on the Indian Ocean islands, which are occupied by six species, and in Africa with four mainland species (Eurasian Scops Owl Otus scops, African Scops-Owl Otus senegalensis, Sokoke Scops Owl Otus ireneae, Sandy Scops Owl Otus icterorhynchus) and two insular species (São Tomé Scops Owl Otus hartlaubi, Pemba Scops Owl Otus pembaensis) (Marks et al. 1999).
Fuchs et al. (2008) suggested that the rapid divergence following the initial colonization of the Indian Ocean islands (Madagascar and the Comoros archipelago) led to distinctive genetic lineages, and this result has favoured the recognition of each insular Otus taxon as a distinct species (Fig. 1). Intriguingly, molecular DNA sequence data suggest that several species have strikingly different affinities than recognized under traditional hypotheses, suggesting that morphology and vocalizations are weak characters with which to define phylogenetic relationships in scops owls. For example, O. pembaensis proved not to be conspecific with Madagascar Scops Owl Otus rutilus, contrary to previous hypotheses (Marks et al. 1999), but instead is closely related to mainland O. senegalensis (Fuchs et al. 2008). Likewise, the São Tomé endemic Otus hartlaubi was found to have strong affinities with O. senegalensis. Even more surprisingly, the Seychelles Scops Owl Otus insularis formed a well-supported clade with Oriental Scops Owl Otus sunia (Fuchs et al. 2008).
Samples of Socotra Scops Owl, currently considered to be a subspecies of O. senegalensis, were unavailable to Fuchs et al. (2008), and the same was true of several taxa found on the Arabian Peninsula (Pallid Scops Owl Otus brucei, and Otus senegalensis pamelae) and East Africa (northern Somalia, Ethiopia, Kenya), where several subspecies of O. senegalensis have been described on the basis of differences in voice, plumage coloration and size (Chapin 1930, Meinertzhagen 1948, Keith & Twomey 1968) but are not generally recognized in the modern literature (Kemp 1988, Marks et al. 1999, Dickinson 2003, König et al. 2008). The affinities of these taxa are still poorly known and this uncertainty is reflected by frequent changes in their taxonomic treatment (Marks et al. 1999, König et al. 2008). Since Sclater (1924), the Socotran population has usually been treated as a subspecies of O. senegalensis (Otus senegalensis socotranus), but it is sometimes placed together with Pallid Scops Owl O. brucei (e.g. Kemp 1988, König et al. 1999), whose range otherwise encompasses the eastern part of the Arabian Peninsula extending east to western Pakistan and north to southeastern Turkey and Central Asia (Marks et al. 1999). This taxon's treatment as a subspecies of O. senegalensis appears to have been influenced by the report ‘that the song on Socotra is identical to that in Kenya’ (A. Forbes-Watson in Marshall 1978). However, recently König et al. (2008) and Porter and Aspinall (2010) considered socotranus to represent a separate species.
Similarly, scops owls from southern Arabia, currently recognized as Otus senegalensis pamelae, have also been suggested to be closely related to O. brucei (Marks et al. 1999), although Marshall (1978) already noted that this taxon's vocalizations are ‘intermediate between the stutter of Otus senegalensis senegalensis and the purr of O. sunia distans of Thailand.’ Furthermore, the systematic affinities of O. brucei remain unclear. It has been treated either as a subspecies of Otus scops (Meinertzhagen 1948) or Otus sunia prior to vocal studies that supported its specific status (Marshall 1978, Roberts & King 1986).
We here examine further the patterns of phenotypic differentiation among scops owl taxa, and discuss these with respect to phylogenetic relationship highlighted by our analyses of mitochondrial and nuclear sequence data.
Methods
We collected blood samples for the Socotra Scops Owl and used toe-pads from museum specimens held at the Natural History Museum, Tring, UK, for O. brucei, O. s. pamelae and O. s. senegalensis collected in Ethiopia, Somalia and Kenya. In addition, our taxonomic sampling was completed with several species included in Fuchs et al. (2008; Table 1).
| Species | Voucher/Tissue number | Geographical origin | Myoglobin | TGFB2 | ND2 | ND3 |
|---|---|---|---|---|---|---|
| Bubo bubo | MNHN 24-55 | France | EU601069 | EU600949 | EU601029 | EU600992 |
| Otus capnodes | MNHN 30-10J | Anjouan | EU601078 | EU600957 | EU601038 | EU601000 |
| Otus hartlaubi | MNHH 32-04G | São Tomé | EU601072 | EU600952 | EU601032 | EU600995 |
| Otus insularis | D. Currie 5H21863 | Mahé, Seychelles | EU601059 | EU600940 | EU601022 | EU600983 |
| Otus insularis | D. Currie 5H21866 | Mahé, Seychelles | EU601060 | EU600941 | EU601023 | EU600984 |
| Otus ireneae | MNHN 32-06J (M. Virani C30577) | Kenya | EU601077 | EU600956 | EU601037 | EU600999 |
| Otus lettia lettia | MNHN JF142 | Laos | EU601073 | EU600953 | EU601033 | EU600996 |
| Otus lettia ussuriensis | UWBM 75379 | Russia | EU601075 | EU600955 | EU601035 | EU600998 |
| Otus longicornis | FMNH 433020 | Luzon, Philippines | EU601084 | EU600952 | EU601043 | EU601005 |
| Otus longicornis | ZMUC 114206 | Isabela, Philippines | EU601063 | No sequence | EU6010526 | EU600987 |
| Otus mayottensis | MNHN R22 | Mayotte | EU601087 | EU600965 | EU601046 | EU601008 |
| Otus megalotis | FMNH 433019 | Luzon, Philippines | EU601083 | EU600961 | EU601041 | EU601004 |
| Otus megalotis | ZMUC 114208 | Isabela, Philippines | EU601064 | EU600944 | EU601027 | EU600988 |
| Otus mirus | FMNH 357429 | Mindanao, Philippines | EU601099 | EU600978 | EU601057 | EU601020 |
| Otus moheliensis | MNHN E-135 | Mohéli | EU601086 | EU600964 | EU601045 | EU601007 |
| Otus pauliani | MNHN R24 | Grande Comore | EU601100 | EU600979 | EU601058 | EU601021 |
| Otus pembaensis | MNHN uncatalogued | Pemba Island | EU601090 | EU600967 | EU601048 | EU601010 |
| Otus pembaensis | MNHN uncatalogued | Pemba Island | EU601091 | EU600968 | EU601049 | EU601011 |
| Otus madagascariensis | FMNH 393149 | Madagascar | EU601082 | EU600960 | EF198290 | EU601003 |
| Otus rutilus | FMNH 431150 | Madagascar | EU601068 | EU600948 | EF198307 | EU600991 |
| Otus scops | MNHN 23-5F | France | EU601079 | EU600958 | EU601039 | EU601001 |
| Otus senegalensis | MVZ uncatalogued | South Africa | EU601098 | EU600976, EU600977 | EU601053 | EU601019 |
| Otus spilocephalus | MNHN 15-58 | China | EU601080 | EU600980 | EU601040 | No sequence |
| Otus sunia | MNHN 6-98 | Thailand | EU601081 | EU600959 | EU601041 | EU601002 |
| Strix aluco | MNHN 24-27 | France | EU601070 | EU600950 | EU601030 | EU600993 |
| New sequences | ||||||
| Otus brucei | BMNH, 1979.2.22 (T) | Oman | KC138811 | No sequence | KC138817 | KC138825 |
| Otus brucei | BMNH, 1979.11.06 (T) | Oman | KC138811 | No sequence | KC138818 | KC138826 |
| Otus s. pamelae | BMNH, 1977.21.11 (T) | Saudi Arabia | KC138812 | No sequence | KC138819 | KC138827 |
| Otus s. pamelae | BMNH, 1937.4.17 (T) | Saudi Arabia | KC138812 | No sequence | KC138820 | KC138828 |
| Otus s. senegalensis | BMNH, 1912.12.23.44 (T) | Orr Valley, Kenya | KC138813 | No sequence | KC138821 | KC138829 |
| Otus s. senegalensis | BMNH, 1965.M.5013 (T) | North Somalia | KC138814 | No sequence | KC138822 | KC138830 |
| Otus s. senegalensis | BMNH, 1923.8.7.7553 (T) | North Somalia | KC138815 | No sequence | KC138822 | KC138830 |
| Otus s. senegalensis | BMNH, 1887.11.11.44 (T) | Abyssinia, Ethiopia | No sequence | No sequence | KC138823 | KC138831 |
| Otus s. socotranus | MNHN Uncatalogued (B) | Mer Koh, Socotra Island | KC138816 | KC138810 | KC138824 | KC138832 |
| Otus s. socotranus | MNHN Uncatalogued (B) | Mer Koh, Socotra Island | KC138816 | KC138810 | KC138824 | KC138832 |
| Otus s. socotranus | MNHN Uncatalogued (B) | Mer Koh, Socotra Island | KC138816 | KC138810 | KC138824 | KC138832 |
- BMNH, Natural History Museum, Tring; FMNH, Field Museum of Natural History, Chicago; MNHN, Muséum National d'Histoire Naturelle, Paris; MVZ, Museum of Vertebrate Zoology, Berkeley; UWBM, University of Washington, Burke Museum, Seattle; ZMUC, Zoological Museum, University of Copenhagen.
Laboratory procedures
Total genomic DNA was extracted from blood preserved in EDTA using a CTAB-based protocol (Winnepenninckx et al. 1993). DNA was extracted from toe-pads using a DNeasy Tissue Kit (Qiagen Inc., Valencia, CA, USA) adding 20 μL of dithiothreitol (DTT; 0.1 m). Genomic extractions of toe-pads were performed separately from the DNA extractions of fresh tissues of Socotra Scops Owl. Prior to each extraction, the workstation and all equipment were cleaned using a bleach solution and UV light was used to help prevent contamination.
Two mitochondrial markers, nicotinamide dehydrogenase subunits 2 and 3 (ND2, ND3), and two nuclear introns (myoglobin intron-2 (MB) and TGFb2 intron-5 (TGFb2)) were amplified via the polymerase chain reaction (PCR) and sequenced. ND2 and ND3 were PCR-amplified and sequenced using primer pairs L5219/H6313 (Sorenson et al. 1999) and L10755/H11151 (Chesser 1999), respectively. The MB nuclear intron was PCR-amplified and sequenced using MYO2/MYO3F (Slade et al. 1993, Heslewood et al. 1998). TGFb2 was PCR-amplified and sequenced using primer pairs TG5/TG6 (Bures et al. 2002). The following internal primers were designed to amplify and sequence DNA obtained from the toe-pads: ND2intL1 5′-CATYAARTRYTTCCTAGTACAAGC-3′; ND2intL2 5′-CAGCYATTGCAATAAAACTAGGAC-3′; ND2intR1 5′-GGACAATGRGACATCACACAA -3′; ND2intR2 5′-CCTAAAYCAAACACAAATCCGA-3′; ND3intL 5′-CTVCCATTCTCARTMCGATTCTTC-3′; ND3intR 5′-CCTATTCCTYCTRTTY GAYCTRG-3′; MYO2intL 5′-TTATTTACTGWTGGCTAGTTGG-3′; MYO2intR 5′-GGTATGTGAATATCCAAGTTTAGA-3′. We obtained DNA sequences of 500 and 320 bp from toe-pad samples for ND2 and MB, respectively. To enable detection of possible contamination, negative controls were included during PCR-amplification of DNA extracts, which utilized the Illustra Hot Start Mix RTG (Buckinghamshire, UK).
Determining the phase of alleles
We used Phase v2.1.1 (Stephens et al. 2001, Stephens & Donnelly 2003) to infer the association among heterozygous sites for each nuclear locus and individual. Three runs using different seed values were performed and results were compared across runs. We used the recombination model and ran the iterations of the final run 10 times longer than for the other two runs. The best haplotype estimate was used for the phylogenetic analyses.
Phylogenetic analyses
Phylogenetic analyses including estimation of individual gene trees and a concatenated approach were conducted using maximum likelihood (ML) and Bayesian inference (BI), as implemented in RAxML v7.0.4 (Stamatakis 2006, Stamatakis et al. 2008) and MrBayes 3.1.2 (Ronquist & Huelsenbeck 2003), respectively. The most appropriate models of nucleotide substitution were determined using TOPALi v2.5 (Milne et al. 2009) and the Bayesian information criterion (BIC). Maximum likelihood and Bayesian analyses under the concatenated approach were performed, allowing substitution model parameters to differ among loci (Nylander et al. 2004). For the gene tree analyses, we ran several preliminary analyses by changing the branch-length prior, from unconstrained:exp(10) to unconstrained:exp(200), and the heating temperature from 0.2 to 0.1. Better mixing was always achieved using a temperature of 0.1 instead of the 0.2 default value. The best-fit brlens prior was selected using the Bayes factor (BF; Nylander et al. 2004). A value > 4.6 for lnBF was considered to be strong evidence against the simpler model (Jeffreys 1961). For MrBayes 3.1.2, we used default priors for the base frequency and substitutions models. For the gene tree analyses, we observed better mixing of the chains and higher likelihood using the unconstrained:exp(10) branch-length prior for the mtDNA data and unconstrained:exp(150) or unconstrained:exp(200) for nuclear data. Analyses of the concatenated data were performed using an unconstrained:exp(100) prior for estimation of the branch-lengths. In all MrBayes analyses, four Metropolis-coupled Markov chain Monte Carlo runs, one cold and three heated, were conducted for 20 million iterations each, with trees sampled every 1000 iterations. Two independent Bayesian runs initiated from random starting trees were performed for each dataset, and the log-likelihood values and posterior probabilities were checked to ascertain that the chains had reached stationarity. We ensured that the potential scale reduction factor (PSRF) approached 1.0 for all parameters and that the average standard deviation of split frequencies converged towards zero. For the concatenated data, we use the consensus sequences from the two alleles for the two nuclear loci, using the IUPAC code for the heterozygous sites. The two independent Bayesian runs were combined to estimate the consensus topology and posterior probabilities of clades.
We used Tracer v1.5 (Rambaut & Drummond 2007) to determine that our sampling of the posterior distribution had reached a sufficient effective sample size (ESS > 200) for meaningful parameter estimation.
Genetic distance
Uncorrected-p genetic distances among taxa were calculated using the concatenated mitochondrial sequences (ND2 and ND3). According to evolutionary rates estimated from complete mitochondrial genomes for Hawaiian honeycreepers (Lerner et al. 2011), the ‘standard’ cytochrome-b gene evolves at a slower rate (0.014 substitutions per site per million years) than ND2 (0.029 s/s/my) and ND3 (0.024 s/s/my) used in the present study to calculate genetic distances among taxa.
Morphological data
For museum specimens of O. senegalensis, O. s. pamelae, O. s. socotranus and O. sunia (Supporting Information Table S1), we obtained the following biometric measurements: wing length (flattened) and tail length, using a standard metal wing-rule with a perpendicular stop at zero (measured to 0.5 mm), and culmen length (to skull), culmen width (at the feathers) and tarsus length (measured from the tarsal joint to the last complete scute before the toes diverge), using digital callipers (to a precision of 0.01 mm). All measurements were taken by G.M.K.
All measurements were log-transformed to normalize the distribution and to allow for allometry. However, morphological variables were not size-standardized because size is an important taxonomic criterion (Helbig 2000, Kaboli et al. 2007) and must be considered with shape in studies of morphological characters (Mosimann & James 1979). Sexual dimorphism was tested using analysis of variance (anova). Principal component analyses (PCA) using correlation matrices were performed with five biometric variables to visualize patterns of covariation in a multivariate space. In addition, discriminant function analyses (DFA) were performed using a priori defined groups. Groups included in each DFA were chosen on the basis of our phylogenetic results: O. s. socotranus was compared with its closest relatives (O. sunia, O. insularis) and O. s. pamelae was compared with five O. senegalensis populations from eastern Africa. All statistical analyses were conducted using Statistica version 6 (StatSoft Inc, Tulsa, OK, USA).
Vocalization
We qualitatively compared sound recordings (n = 38) of O. senegalensis, O. s. pamelae and O. insularis. Localities and sample sizes are presented in Supporting Information Table S2. Recordings were obtained from two online databases, Xeno-canto (XC; www.xeno-canto.org) and the Cornell Lab of Ornithology Macaulay Library (ML; http://macaulaylibrary.com/), commercially available recordings (Chappuis 2000), from colleagues or from personal recordings. Sonograms were created using WaveSurfer 1.8.8p4 (http://sourceforge.net/projects/wavesurfer/) under the following parameters: FFT window length: 2048 points; window type: Hamming; analysis bandwidth: 57 Hz; and window length: 768 points. Vocalizations are commented upon in the Discussion with respect to phylogenetic relationships.
Results
Phylogeny
The length of the alignment, number of individuals sequenced, DNA substitution models, likelihood scores and clock model selected for each individual locus are indicated in Table 2.
| Locus | ND2 | ND2 | ND3 | mtDNA | MB | TGFb2 | Concatenated |
|---|---|---|---|---|---|---|---|
| Length (bp) | 1041 | 500 | 351 | 1392 | 723 | 593 | 2708 |
| No. of individuals (haplotypes) | 34 (29) | 34 (29) | 34 (29) | 34 (30) | 33 (26) | 27 (33) | 32 |
| Substitution model | TrN+Γ | HKY +Γ | K81uf +Γ +I | partitioned | TrnEf | K81 + Γ | partitioned |
| ML score | −6208.53 | −2958.01 | −2066.58 | −8276.17 | −1459.86 | −1474.96 | −10943.43 |
| BI harmonic mean | −6256.55 | −3019.23 | −2138.1 | −8351.17 | −1514.25 | −1565.65 | −11098.72 |
Total evidence
The analyses of the concatenated dataset resulted in a 50% majority consensus rule tree that was very similar to the mtDNA topology. However, most nodes received better support values (Fig. 2). The African species Otus ireneae, endemic to the coastal forests of Kenya and Tanzania, was found to be the sister-taxon to all of the remaining Otus scops owls (Fig. 2). The phylogenetic tree was then divided into two clades. One Indo-Malayan clade included Mountain Scops Owl Otus spilocephalus, Otus lettia and Philippine Scops Owl Otus megalotis, whereas the other clade consisted of all the western Indian Ocean taxa as well as O. scops and all African mainland and insular taxa. This clade can be divided further into two highly supported sub-clades, which comprised Indo-Malayan/Indian Ocean taxa and Afro-Palaearctic taxa (Fig. 2).
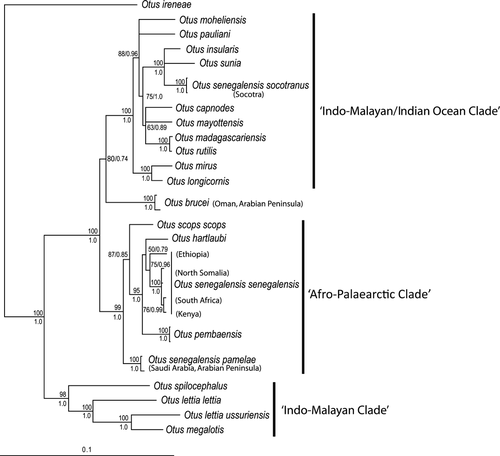
The two Arabian taxa belonged to different clades: O. brucei was recovered with moderate support to be the first offshoot in the Indo-Malayan/Indian Ocean (ML = 80, BI = 0.74), whereas O. s. pamelae grouped with the Afro-Palaearctic taxa in the second clade. Socotra Scops Owl O. s. socotranus grouped with Oriental Scops Owl and Seychelles Scops Owl (ML = 100, BI = 1.0), but the relationships among these three species were not resolved. The latter clade was closely related to the other Indian Ocean taxa (ML = 88, BI = 0.96). Two species endemic to the Philippines (Luzon Scops Owl Otus longicornis and Mindanao Scops Owl Otus mirus) were the sister-group to this mostly Indo-Malayan clade (ML = 100, BI = 1.0). The African Scops Owl was not monophyletic because the Arabian taxon O. s. pamelae was sister to the clade formed by O. scops, O. s. senegalensis, O. hartlaubi and O. pembaensis (ML = 99, BI = 1.0). There was substantial mitochondrial genetic divergence among samples of O. s. senegalensis; the individual collected in Ethiopia differed by up to 3.2% from other O. s. senegalensis individuals included in our study.
mtDNA
The final mtDNA data retained for the analyses included 30 individuals and 1392 bp (ND2: 1041 bp, ND3: 351 bp). Four individuals were not included in the analyses because they had the same haplotype as another conspecific individual (the two Seychelles Scops Owls, the three Socotra Scops Owls and the two Pemba Scops Owls). The tree resulting from the concatenated mtDNA data had uneven support, with most of the relationships among species of recent origin having low support (BI = 0.50) whereas the backbone relationships of the Otus clade often received strong support (ML = 100, BI = 1.0) (Supporting Information Fig. S1).
Nuclear data
The two nuclear loci were phased prior to the phylogenetic analyses and only unique alleles were retained for tree reconstruction. Sharing of allele sequences was widespread among closely related species. Three alleles were shared among species for MB, the first being found among several members of the Indo-Malayan/Indian Ocean clade (O. insularis, O. mirus, O. rutilus, O. longicornis, Karthala Scops Owl O. pauliani and O. s. socotranus). The second and third alleles were shared among species of the Afro-Palaearctic clade (O. pembaensis, O. s. pamelae and O. s. senegalensis; and O. scops and O. s. senegalensis, respectively). For TGFb2, alleles were only shared between O. rutilus and Mayotte Scops Owl Otus mayottensis. Overall, there were a greater number of alleles for TGFb2 than for MB (33 vs. 26).
The tree resulting from the BI of the MB alleles recovered the same five primary lineages highlighted by the mtDNA (O. irenenae, O. brucei, Indo-Malayan/Indian Ocean clade, Afro-Palaearctic clade and Indo-Malayan clade) but relationships among these five lineages were not supported. Relationships within these clades formed a polytomy (Supporting Information Fig. S2).
The TGFb2 tree exhibited greater structure. Otus ireneae diverged at the base of the Otus clade (ML = 80, BI = 1.0) and members of the ‘Indo-Malayan clade’ were paraphyletic with respect to the ‘Indo-Malayan/Indian Ocean’ and ‘Afro-Palaearctic’ clades. Members of the two latter clades were recovered in a general polytomy (Supporting Information Fig. S3). Yet, as for other loci, O. s. socotranus clustered with O. sunia and O. insularis (ML = 43, BI = 0.93) whereas O. s. senegalensis clustered with O. pembaensis (ML = 75, BI = 0.96). Sequences from O. s. pamelae and O. brucei could not be obtained for this locus.
The concatenation of the nuclear data using unphased sequences yielded a 50% majority rule consensus tree that was very similar to the MB and TGFb2 gene trees. A major discrepancy involved the relationships of Pallid Scops Owl, which clustered with Sokoke Scops Owl at the base of the Otus clade (ML = 65, PP = 0.89), a result that was not recovered in any of the single gene tree analyses. We attribute this result to the joint effect of MB having limited variation to resolve its relationships and the fact that we could not sequence TGFb2 for O. brucei. The three primary clades within Otus (Indo-Malayan, Afro-Palaearctic, Indian Ocean/Indo-Malayan) were all recovered as monophyletic, although support was variable (ML = 58, BI = 0.93; ML = 62, BI = 0.94; and ML = 90, BI = 1.0, respectively), but no resolution was achieved within each of the clades.
Vocalization
Otus s. socotranus
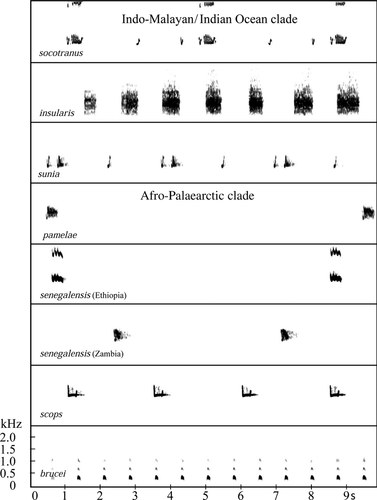
Although our sample size was of only three different recordings, G.M.K. and R.F.P. have considerable field experience with O. s. socotranus and its vocalizations (G.M.K. during 1 week of intensive general avifaunal fieldwork in spring 1993 and R.F.P. during 11 visits to the island between 1993 and 2011, for a total of 6 months). R.F.P. has heard a total of 68 different socotranus individuals singing as part of focused fieldwork on the taxon designed to elucidate its general biology, natural history and population size. The song is a repeated, low-pitched woop woop da-pwoorp…woop woop da-pwoorp…woop woop da-pwoorp, each phrase lasting c. 2 s with similar-length intervals between them (Supporting Information Audio File S1). The two woop notes are much less audible, except at close range. This is very similar to at least some songs of O. sunia, but less so to O. senegalensis over most of mainland Africa. We found no evidence of close vocal similarities between O. s. socotranus and other populations of O. senegalensis, including those in Arabia. Across much of eastern and southern mainland Africa, senegalensis gives a soft, purring trill, kjurrrr, lasting up to 1 s (but often just 0.5 s). Inter-phrase intervals are typically 4–8 s. Sonograms of typical songs are presented in Figure 3.
Otus pamelae
Southern Arabian O. s. pamelae differs from other populations of senegalensis. The sound is obviously higher pitched than that of eastern and southern African populations, each phrase lasts c. 1 s, and the notes are harsher sounding, with a more noticeable tremolo. By contrast, inter-phrase intervals are similar to those in eastern and southern African populations. The sole recording available to us from the Republic of Somaliland appears to present a similar vocal type to those in southern Arabia and is the focus of ongoing research (G. M. Kirwan & R. F. Porter unpubl. data). We do not agree with Marshall (1978) that pamelae is ‘intermediate’ between senegalensis and sunia in its song; it is clearly similar to the former in most respects.
Morphology
We did not observe significant sexual dimorphism in morphometric traits (anova, all comparisons P > 0.05, results not shown) except for the bill length of socotranus (F1,17 = 13.92, P = 0.0001). All individuals were thus pooled to perform the multivariate analyses. Descriptive statistics for each taxon are shown in Table 3.
| Afro-Palaearctic clade | Indo-Malayan/Indian Ocean clade | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Otus senegalensis pamelae | Otus senegalensis senegalensis | Otus senegalensis socotranus | Otus sunia | Otus insularis | |||||
| Arabian Peninsula | Somalia | Ethiopia | Kenya | Sudan | Uganda | Socotra | India | Seychelles | |
| Wing length | 7, 137.6 (5.6) | 5, 121.4 (2.9) | 3, 130.3 (3.5) | 7, 132.1 (6.8) | 7,134.1 (4.8) | 6, 131.7 (6.1) | 21, 129.3 (3.8) | 17, 145.7 (3.8) | 3, 164.7 (5.9) |
| Tail length | 7, 60.2 (3.4) | 5, 51.8 (0.4) | 3, 57 (3) | 7, 57.2 (4.1) | 7, 60.1 (3.4) | 6, 60.2 (4) | 20, 54.1 (4.3) | 17, 64.9 (3.3) | 3, 77 (4.3) |
| Tarsus length | 7, 30.3 (1.9) | 4, 22.7 (1.5) | 3, 24.3 (1) | 7, 24.5 (1.48) | 6, 25 (2.1) | 6, 25 (1.7) | 13, 24.4 (2.3) | 9, 25 (2) | 3, 32.1 (0.7) |
| Bill length | 7, 18.4 (1.4) | 5, 16.9 (0.6) | 3, 17.6 (0.6) | 7, 18.3 (0.8) | 7, 17.4 (1.2) | 6, 16.7 (1.3) | 21, 19.4 (0.9) | 16, 18.5 (1.1) | 3, 23.5 (1.3) |
| Bill width | 7, 7.6 (0.5) | 5, 7.5 (0.7) | 3, 7.4 (0.6) | 7, 8.3 (0.4) | 7, 7.8 (0.4) | 6, 7 (0.5) | 13, 9.1 (0.8) | 16, 6.7 (0.7) | 3, 9.9 (1.3) |
PCA
Table 4 summarizes the eigenvalues, the proportion of variance captured by the first two PCs and factor loadings for each biometric variable and each PCA analysis. The first two components (PC1, PC2) together accounted for a large amount of the variance (> 70%).
| A – PC1 | A – PC2 | B – PC1 | B – PC2 | |
|---|---|---|---|---|
| Eigenvalue | 2.73 | 1.58 | 2.47 | 1.39 |
| Variance explained | 54.54% | 31.54% | 49.41% | 27.94% |
| Factor loadings | ||||
| Wing | 0.88 | −0.37 | 0.90 | −0.13 |
| Tail | 0.82 | −0.46 | 0.86 | −0.34 |
| Tarsus | 0.84 | 0.20 | 0.79 | −0.14 |
| Bill length | 0.74 | 0.59 | 0.51 | 0.68 |
| Bill width | 0.12 | 0.91 | 0.20 | 0.87 |
Morphological divergence of O. s. socotranus. PC1 explained 54.5% of the variation and described size differences among taxa, each standardized biometric variable, except bill width, being roughly equally and positively correlated with PC1. PC2 captured a comparatively minor amount of variation (31.5%) and described differences in shape among taxa, bill width having the highest factor loading. There was clear separation of PC1 and PC2 scores among socotranus, insularis and sunia individuals (anova PC1, F2,22 = 101.46, P < 0.0001; anova PC2, F2,22 = 31.28, P < 0.0001). Socotra Scops Owls had shorter wings and a shorter tail than O. insularis. In addition, PC2 clearly separated O. sunia from socotranus, which possessed a relatively larger and wider bill. PC3 (9% of the total variation, F2,22 = 0.17, P = 0.84) and PC4 (4% of the total variation, F2,22 = 0.29, P = 0.75) did not provide any clear separation among these three taxa.
Morphological divergence of O. s. pamelae. As for the previous PCA, each standardized morphometric variable was positively correlated with PC1 and contributed roughly equally to PC1, which explained most of the variation (49.4%). PC2 captured 27.94% of the variance and was positively correlated with bill length and bill width. Otus s. pamelae was partially separated from its African counterparts along PC1 (anova PC1, F5,27 = 6.74, P = 0.0003). Most O. s. pamelae individuals tended to be larger in wing and tail length than their African counterparts. There was no clear separation of PC2 scores among scops owls whatever their geographical origin. PC3 and PC4, which explained 12 and 8% of the total variation, respectively, did not permit further separation of O. s. pamelae from African taxa (PC3, F5,27 = 1.66, P = 0.18; PC4, F5,27 = 1.38, P = 0.36).
DFA
Morphological divergence of O. s. socotranus (Fig. 4). A multivariate anova was strongly significant (Wilks' lambda = 0.011, F10,36 = 30.1, P < 0.0001) and fully separated the three taxa. The variables that contributed most to the discrimination between groups are wing length (root 1: standardized coefficient = 1.06) and bill length (root 2: standardized coefficient = −0.71). Socotran birds were shorter in wing length than sunia and insularis, and had a wider and larger bill than sunia. The classified matrix derived from the DFA correctly classified 100% of socotranus (n = 7) with high posterior probabilities (PP = 1.0).
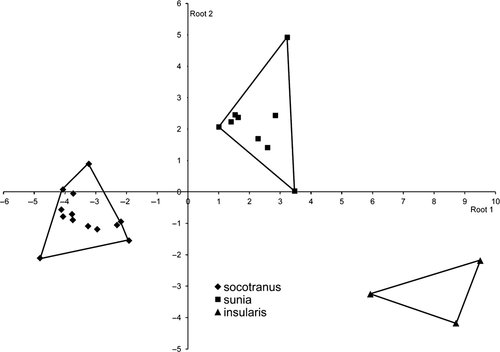
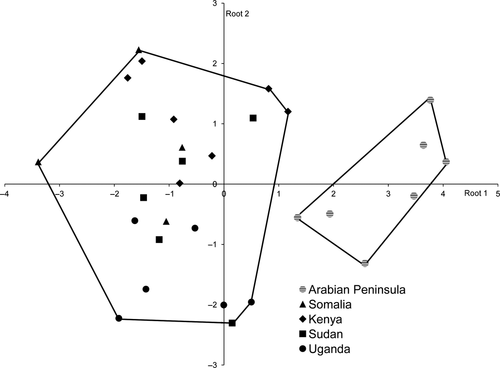
Morphological divergence of O. s. pamelae (Fig. 5). The first two discriminant functions explained 87% of the variability in the dataset. DFA significantly differentiated biometric characteristics among groups (manova, Wilks' lambda = 0.080, F25,86 = 3.50, P < 0.0001). However, there was no clear pattern of geographical structure among mainland African Scops Owls. In contrast, O. s. pamelae was clearly separated from the other groups: O. s. pamelae is larger in wing and tarsus length than its African counterparts. The classified matrix derived from the DFA correctly classified 100% of O. s. pamelae with high posterior probabilities (PP > 0.99) for five individuals and low posterior probabilities for only two individuals (PP < 0.50).
Discussion
Species limits
We recovered the same general topology as that of Fuchs et al. (2008), which was expected given that the taxonomic and gene sampling was very similar, with only a few nodes showing less support. Otus brucei was recovered with moderate support to occupy a basal position within the Indo-Malayan sub-clade, whereas all senegalensis taxa belonged to the Afro-Palaearctic sub-clade.
The Pallid Scops Owl is considered by some authorities to be a subspecies of either O. scops or O. sunia, or regarded as distinct, forming a superspecies with O. senegalensis, O. scops and O. sunia (Marks et al. 1999). Our molecular results do not support such systematic arrangements among these four species. Indeed, the Pallid Scops Owl is not closely related to either O. scops or O. senegalensis, but appears to be the most ancient lineage that diverged from the Indo-Malayan and Comorian sub-clade (Fig. 1). Otus brucei is in widespread sympatry with O. scops without evidence of interbreeding, and also exhibits unique vocal (for sonograms see Rasmussen & Anderton 2005: 238) and morphological characters (Shirihai 1993) that fully support specific taxonomic treatment according to the guidelines proposed by Helbig et al. (2002).
The Socotra Scops Owl is currently treated as a subspecies of O. senegalensis in most of the literature (e.g. Marks et al. 1999, Dickinson 2003, Clements 2007). This taxonomic treatment is not supported by our molecular results and our qualitative analyses of vocalization data. Together with O. insularis and O. sunia, the Socotra Scops Owl forms a well-supported monophyletic group nested within the Indo-Malayan/Indian Ocean clade. Socotra Scops Owl vocalizations are reminiscent of O. sunia, in accordance with the close relationship to the latter taxon highlighted in our phylogenetic tree.
Mitochondrial uncorrected-p distances from its sister species O. sunia and O. insularis are 4.7 and 3.5%, respectively, indicating that the two taxa are clearly genetically distinct. These genetic distances based on ND2 and ND3 genes are of the same order of magnitude as genetic distances usually found in the cytochrome-b gene between sister-species of raptors (Wink & Sauer-Gürth 2000). Socotra Scops Owl is clearly distinguishable morphologically from its closest relatives; it is significantly smaller than O. sunia and O. insularis in wing and tail length. The Socotra Scops Owl is further differentiated from O. sunia with respect to its larger bill length and bill width. Otus senegalensis socotranus is definitively monomorphic (occurring solely in a grey-brown morph; G.M. Kirwan & R.F. Porter pers. obs.), unlike O. sunia, which possess two colour morphs (rufous and grey-brown) in most or all populations, even for most or all insular subspecies, e.g. Otus sunia japonicus (Japan) and Otus sunia leggei (Sri Lanka), as well as mainland taxa, e.g. O. s. sunia (northern Indian subcontinent) and Otus s. stictonotus (Far East Asia) (G.M. Kirwan pers. obs.; Rasmussen & Anderton 2005, König et al. 2008). Overall, our results suggest that this insular taxon represents an independent evolutionary lineage. Socotra Scops Owl possesses several independent diagnostic characters (sensu Helbig et al. 2002) that strongly favour its status as a species Otus socotranus, supporting the treatment adopted by König et al. (2008), Jennings (2010) and Porter and Aspinall (2010).
Another taxon that deserves further attention is O s. pamelae and its position within the Afro-Palaearctic clade (ML = 99, BI = 1.0). This southern Arabian taxon is highly divergent from African senegalensis (uncorrected-p mitochondrial genetic distance = 4%), making O. senegalensis, as currently defined, polyphyletic. The song of pamelae is very different from that of O. scops and O. brucei but more similar to that of O. senegalensis. It nevertheless differs from the latter's song in being higher pitched, sounding ‘scratchier’ and having more prolonged notes; the song sounds two-parted, due to the much quieter first note (G.M. Kirwan & R.F. Porter pers. obs., König et al. 2008). In terms of biometrics, our results clearly suggest that pamelae is longer winged and longer legged than mainland African populations of senegalensis. In comparison with populations of O. senegalensis in continental Africa, Arabian pamelae is distinguished in being paler overall, with less distinct streaking over the underparts and a less obvious whitish line on the scapulars (König et al. 2008). Such plumage characteristics might result from adaptation to arid habitats, as has been demonstrated for other desert-dwelling species (Guillaumet et al. 2008). Our study reveals that Arabian Scops Owls possess several diagnostic genetic and phenotypic characters. We therefore consider the most appropriate taxonomic treatment is to recognize Arabian Scops Owl as a species, Otus pamelae, and not as a subspecies of O. senegalensis as it was originally described based solely on morphological data (Bates 1937).
Among O. s. senegalensis (sensu stricto), the Ethiopian lineage is genetically distinct (uncorrected-p mitochondrial distance = 3.2%) from other lineages found in eastern (northern Somalia and Kenya) Africa and in South Africa. We included only three Ethiopian scops owls in our biometric analyses and these were not differentiated from their African counterparts. The level of phenotypic distinctness of the Ethiopian population remains to be established with a larger sample size. Furthermore, as our phylogenetic analyses only included one individual, collected in the late 19th century without precise data concerning its geographical provenance, we advocate additional genetic studies prior to any taxonomic change.
The two Somaliland scops owls sampled were genetically poorly differentiated from their Kenyan and South African counterparts. However, populations from the highlands of Somaliland are vocally distinct from other eastern African populations and possess a similar song to the phylogenetically distantly related Arabian O. pamelae. In accordance with Fuchs et al. (2008) concerning the affinities of the Indian Ocean scops owls, the present study affirmed that vocal differences among Otus taxa do not always reflect evolutionary affinities among lineages.
Biogeographical aspects
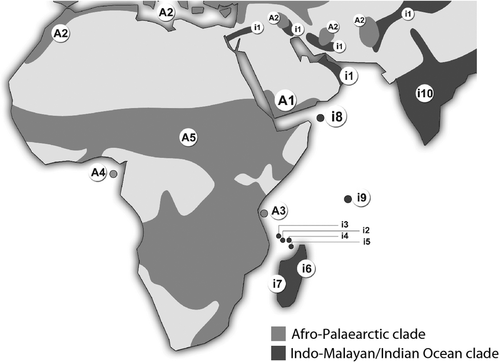
That Otus socotranus is most closely related to Indo-Malayan (O. sunia) and Seychelles (O. insularis) Scops Owls might appear surprising given its geographical location in the Gulf of Aden, closest to the African mainland (Fig. 1). Although Socotra is very distant from other Indian Ocean islands, Socotra Scops Owls are more closely related to other insular Otus taxa than to their Arabian or African counterparts. Surprisingly, of all western Indian Ocean Islands only Pemba Island, located 50 km off mainland Tanzania, was colonized by scops owls from Africa (Fuchs et al. 2008, this study). Having been above sea level consistently since 31 mya (Braithwaite 1987), Socotra has most probably served as a ‘stepping stone’ between Africa and Arabia for several unrelated organisms (Wranik 2003). However, in the case of Otus, the Socotra archipelago represents the westernmost point reached by this Indo-Malayan clade, as it seemingly has never successfully colonized eastern Africa. Furthermore, although usually considered part of the Afrotropical zoogeographical realm, Socotra lies close to this realm's junction with the Palaearctic and Oriental realms, making it unsurprising that flora and fauna representative of both these regions occur on the archipelago (Cheung & DeVantier 2006, Porter & Kirwan 2010). Socotra's modern diversity is the result of complex biogeographical patterns in which vicariant as well as long-distance transoceanic dispersal events have played a major role (Gómez-Díaz et al. 2012). Given its long isolation from the Arabian Peninsula and its ecological characteristics, the archipelago supports high levels of endemism among several zoological and botanical groups, leading to the islands being often referred to as the ‘Galapagos of the Indian Ocean’. For example, among reptiles, 90% of species are endemic (Gómez-Díaz et al. 2012).
The finding that O. socotranus represents a distinctive species brings to 10 the number of avian taxa now recognized at the species level as being endemic to the Socotran archipelago, thereby significantly enhancing its importance as an Endemic Bird Area (Stattersfield et al. 1998). However, further molecular work on endemic Socotran birds is required to elucidate their phylogeography. Although most of these taxa appear to be Afrotropical in origin, the limited available data are as yet largely inconclusive in determining this. For example, Socotra Sunbird Nectarinia balfouri lies outside both clades of Indian Ocean Nectariniidae and taxon sampling was insufficiently detailed to resolve its position and closest relatives (Warren et al. 2003). Furthermore, although the two endemic sparrows, Socotra Sparrow Passer insularis and Abd Al Kuri Sparrow Passer hemileucus have been sampled (Ryan et al. 2010), most of their presumed closest relatives in the Passer motitensis complex (Kirwan 2008) have not. On the other hand, the endemic Socotra Buzzard Buteo socotranus does indeed appear to be most closely related to Afrotropical Buteo (Riesing et al. 2003, Porter & Kirwan 2010).
The two Arabian taxa, O. brucei and O. pamelae, were the first taxa to diverge within their respective Indo-Malayan/Indian Ocean and Afro-Palaearctic clades. This unexpected finding advocates the need for fresh studies to understand the role played by the Arabian Peninsula in the evolution of Otus lineages, and birds in general.
Many studies of mammalian and reptilian taxa with disjunct distributions in the Horn of Africa and southwest Arabia have demonstrated that colonization most frequently occurred from Africa into the Arabian Peninsula (Portik & Papenfuss 2012). Such work has also revealed the role played by the Red Sea as a more or less effective biogeographical barrier. Our results clearly eliminate a possible African origin of Arabian pamelae and underline the important biogeographical role of the Red Sea in preventing any gene flow between African senegalensis and their Arabian counterparts.
Our limited genetic data on the Ethiopian senegalensis lineage imply a long period of genetic isolation from other eastern and southern African Scops Owl populations. Additional studies are clearly needed to investigate relationships among Otus scops owls from the Horn of Africa. However, our preliminary results fully agree with previous observations that the Rift Valley strongly impacted the phylogeographical structure of several species distributed in this region (e.g. Galerida larks, Guillaumet et al. 2008, Ostrich Struthio camelus, Freitag & Robinson 1993, Lanius shrikes, Fuchs et al. 2011). The Horn of Africa is an important hotspot of avian endemism in Africa, with several Endemic Bird Areas having been identified (Stattersfield et al. 1998). Thirty bird species endemic to Ethiopia and Eritrea alone are known to date (Ash & Atkins 2009, Redman et al. 2009) and additional cryptic species probably remain to be identified in this part of Africa.
Acknowledgments
We thank Mark Adams at the Natural History Museum, Tring, for expediting the toe-pad samples of various scops owl populations. For access to relevant specimen material at the same institution, G.M.K. is indebted to Robert Prŷs-Jones and Mark Adams; at the Smithsonian Institution, National Museum of Natural History, in Washington, DC, James P. Dean and Christina A. Gebhard afforded similar assistance, and at the Field Museum of Natural History, Chicago, G.M.K. was assisted by John Bates, Mary Hennen and David Willard. Fieldwork by G.M.K. in Yemen and Socotra was undertaken within the framework of the 1993 Ornithological Society of the Middle East (OSME) South Yemen expedition, while his museum studies in North America were partly supported by a grant from OSME and further enabled by a scholarship award from the Field Museum of Natural History (Chicago). Additional vocal material was kindly supplied by Nik Borrow, Peter Davidson, Jon Hornbuckle, Michael Mills and Dave Showler. Willem-Pier Vellinga (of Xeno-canto) very kindly assisted in preparing the sonograms for this paper. We are also grateful to Céline Bonillo and to the technical staff of the ‘Service de Systématique Moléculaire’ for helping with laboratory work. We thank Daniel Pons who helped to draw Figures 1 and 3 and two anonymous referees, Gary Voelker and Rauri Bowie for helpful comments on an earlier version of the manuscript. This project was supported by the network ‘Bibliothèque du Vivant’ funded by the CNRS, the Muséum National d'Histoire Naturelle, the INRA and the CNS (Centre National de Séquençage).




