The detection of isochromosome i(12p) in malignant germ cell tumours and tumours with somatic malignant transformation by the use of quantitative real-time polymerase chain reaction
Abstract
Aims
Malignant germ cell tumours (GCTs) of the testis are rare neoplasms, but the most common solid malignancies in young men. World Health Organization guidelines divide GCTs into five types, for which numerous immunohistochemical markers allow exact histological subtyping in the majority of cases. In contrast, a germ cell origin is often hard to prove in metastatic GCTs that have developed so-called somatic malignant transformation. A high percentage, up to 89%, of GCTs are characterised by the appearance of isochromosome 12p [i(12p)]. Fluorescence in-situ hybridisation has been the most common diagnostic method for the detection of i(12p) so far, but has the disadvantages of being time-consuming, demanding, and not being a stand-alone method. The aim of the present study was to establish a quantitative real-time polymerase chain reaction assay as an independent method for detecting i(12p) and regional amplifications of the short arm of chromosome 12 by using DNA extracted from formalin-fixed paraffin-embedded tissue.
Methods and results
A cut-off value to distinguish between the presence and absence of i(12p) was established in a control set consisting of 36 tumour-free samples. In a training set of 149 GCT samples, i(12p) was detectable in 133 tumours (89%), but not in 16 tumours (11%). In a test set containing 27 primary and metastatic GCTs, all 16 tumours with metastatic spread and/or somatic malignant transformation were successfully identified by the detection of i(12p).
Conclusion
In summary, the qPCR assay presented here can help to identify, further characterise and assign a large proportion of histologically inconclusive malignancies to a GCT origin.
Introduction
Germ cell tumours (GCTs) encompass a group of neoplasms that develop from primordial germ cells. Whereas ⁓95% of GCTs manifest in the gonads, 5% occur in other regions of the body, such as the retroperitoneum, the mediastinum, or the pineal gland region, resulting in them being divided into gonadal and extragonadal GCTs.1, 2 GCTs represent a heterogeneous group of tumours that are divided into five types according to the World Health Organization, depending on age and sex.3 Type II GCTs not only constitute> 90% of all GCTs in adolescents and adults, but also comprise a variety of distinct histological subtypes. Generally, these tumours are divided into seminomas (SEMs) and non-seminomas (NSEs), which account for 50% and 40% of cases, respectively. The remaining 10% are mixed forms of seminomatous and non-seminomatous tumours.4 NSEs are further subdivided into embryonal carcinomas, teratomas (TEs), choriocarcinomas, yolk sac tumours (YSTs), and NSEs with mixed histological subtypes. Type II GCTs have a common precursor lesion called germ cell neoplasia in situ (GCNIS).5 Regressed seminomas or GCTs with late metastases complete the wide field of possible histological presentations of type II GCT. Numerous immunohistochemical (IHC) markers, such as SALL4, OCT3/4, podoplanin, SOX2, and glypican-3 (GPC3), are available for use in histological subtyping.3 These markers, and especially their combinations, allow the identification of GCT subtypes and establishment of the diagnosis in most cases. However, there are often ambiguous histological and IHC findings. The wide variety of histological subtypes and the frequent occurrence of mixed tumours complicate the assessment of IHC markers.6 Small subsets of GCTs or their metastases develop secondary cancers that resemble ‘somatic’ non-GCT tumours elsewhere in the body, which can be diagnostically challenging.
During the transition from GCNIS to an invasive tumour, neoplastic cells acquire additional genetic material on the p-arm of chromosome 12, usually in the form of isochromosome 12p [i(12p)] or the amplification of specific areas of chromosome 12 (12p gain).7, 8 The term ‘isochromosome’ is used for an unbalanced structural aberration of a chromosome with two nearly identical arms.9 i(12p) was first described in 1983,10 and was subsequently confirmed as a chromosomal tumour marker in GCT. i(12p) has been observed sporadically in other tumours and in Pallister–Killian syndrome. In those few GCTs that lack i(12p), essential chromosomal aberrations affecting the p-arm of chromosome 12 are supposed to induce GCT type II development.11 Consequently, it is of importance to understand which chromosomal changes on the p-arm occur in those GCTs that do not have i(12p).12 Various studies on i(12p)-negative cases have shown that specific sections on the p-arm, such as section 12p11.2–12.1, are overrepresented.13-15 Some of the genes located on chromosome 12 that might play a role in germ cell tumorigenesis are GDF3, DPPA3/STELLA, SOX5, PHC2, and ATF7IP, and proto-oncogenes such as NANOG, CCND2, and KRAS.16 Currently, fluorescence in-situ hybridisation (FISH),11 comparative genomic hybridisation (CGH)13 and polymerase chain reaction (PCR)17 are used for scientific and diagnostic purposes. However, these methods are complex, do not stand alone, or have no defined cut-off values. We therefore sought to elaborate a quantitative real-time PCR (qPCR)-based approach for the detection of i(12p) and the overrepresentation of circumscribed genetic sections on the p-arm of chromosome 12 in i(12p)-negative GCTs by using specific primers and a healthy patient cohort to establish an additional independent molecular method. We have successfully used our method in difficult cases of metastatic tumours of unknown or doubtful origin.
Materials and methods
TISSUE SAMPLES
The tissue samples of 212 patients were collected at the University Medical Centre in Göttingen, Germany and the Institute of Pathology, RWTH Aachen University between 2009 and 2020. The investigated tissues consisted of formalin-fixed paraffin-embedded (FFPE) tissue samples (Table 1). The study was approved by the ethics committees of the medical faculties of Göttingen (no. 18/2/16) and Aachen (EK249/19).
| Patients (n) | 185 |
| Tumours (n) | 149 |
| Histological subtype (n) | |
| SEM | 107 |
| NSE | 26 |
| EC | 7 |
| TE | 3 |
| Mixed | 16 |
| Mixed SEM/NSE | 16 |
| Healthy tissue (control set) | 36 |
- EC, embryonic carcinoma; NSE, non-seminoma; SEM, seminoma; TE, teratoma.
SELECTION OF PRIMER PAIRS
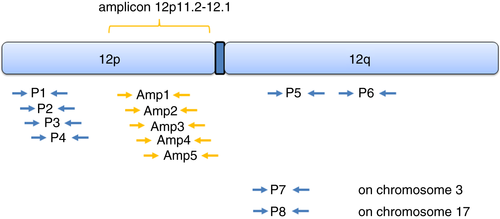
Four primer pairs for the 12p-arm (P1–P4), two primer pairs for the 12q-arm (P5 and P6) and two reference primer pairs for the p-arm of chromosome 3 (P7) and 17q (P8) were defined for the panel (Table 2; Figure 1). In addition, i(12p)-negative cases have shown overrepresentation of small specific regions on the p-arm of chromosome 12, such as 12p11.2–12.1.13-15 For this special purpose, additional primer pairs (Amp1–Amp5) were defined in order to detect a gain of genetic material for this region (Table 3). For amplification of qPCR products, synthetic oligonucleotides from Eurofins MWG Operon (Ebersberg, Germany) and Carl Roth KG (Karlsruhe, Germany) were used. Primer design was performed with the platform primer3web version 4.1.0 (http://bioinfo.ut.ee/primer3/) 18
| Primer | Sequence |
|---|---|
| P1 forward | GGCCTTCTTGCAACATGAGAGTAAG |
| P1 reverse | CAGACTGCACAAAAGGATGGCC |
| P2 forward | GCTCTGTCGGCCTCCATGTCAG |
| P2 reverse | GTCCTCTGTGTGCTCCTCGGC |
| P3 forward | CATGCACTTCCCGCCCTTTTCC |
| P3 reverse | ACTGACAGCTATCTCGCAGACCAAC |
| P4 forward | CTGGGATCTTGGACACTCAGGACAC |
| P4 reverse | TATGTGCCCTTAGACCAGGCAACTG |
| P5 forward | CCTATATCCCCTCTGCCACCAACAC |
| P5 reverse | ACCTCTGCCATGAGAGGCAGTCTTT |
| P6 forward | AGGAAACCTTTGAGAGGCACAGTCG |
| P6 reverse | CCGGGCAATCGCAATAGAGTGTAG |
| P7 forward | CACAGAGTAAAGGCCCCGTGACTTT |
| P7 reverse | ACAGAAGGGCCAGAAAGAACCGAAC |
| P8 forward | TGGGCAGCCCTCATTATCTGGGGCA |
| P8 reverse | ATCCACCCGCCATTGGCATCGAAGC |
| Amp1 forward | GTGGCTCCTCTCCTTTCACACCG |
|---|---|
| Amp1 reverse | TTGTGTACCCTGGAGCCATCGC |
| Amp2 forward | TGTTTGGGTCACAGGGGATGCC |
| Amp2 reverse | TCCTGTGCCAAGTTCCCAACCC |
| Amp3 forward | GAGGTTGGGTCAGACGGTGGTG |
| Amp3 reverse | AATTGCTCCCTCTTTCCCACCCC |
| Amp4 forward | TGCAGTCACTCTACGCAACGCC |
| Amp4 reverse | CCAGCACATCCTCCTCCACGTG |
| Amp5 forward | AGGCCCAATACTCACTGGTCCAC |
| Amp5 reverse | CGGAGGAAAAGAGCAACCCCAAG |
DNA ISOLATION FROM HUMAN FFPE TISSUE AND qPCR
DNA from FFPE tissue was isolated with the innuPREP FFPE DNA Kit-IPC16 (Analytik Jena AG, Jena, Germany) on the InnuPure C16 System (Analytik Jena AG), according to the manufacturer's instructions. The DNA concentration was measured with the NanoDrop 2000 (Thermo Fisher Scientific, Neuendorf, Germany), according to the manufacturer's instructions. For the standard curve, commercially available human genomic DNA (male) was used in a serial dilution: 2.5 ng/µl, 1.25 ng/µl, 0.625 ng/µl, and 0.3215 ng/µl. qPCR was performed with the QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific). The reaction mixture consisted of 2.5 µl of DNA (5 ng/µl), 2.5 µl of primer pair (forward/reverse, 100 pmol/µl), and 5 µl of QuantiNova SYBR Green (QIAGEN GmbH, Hilden, Germany). The following PCR conditions were used: 50°C for 2 min, 95°C for 15 min, and 40 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 1 min. At the end of each PCR run, a melting curve analysis was performed to demonstrate amplification of a specific single PCR product (Figure S1).
DEFINITION OF THE DIFFERENT TEST COHORTS
To establish the qPCR protocol, to evaluate the variance, to calculate standard deviations (SDs), and to define the cut-off values, a control sample set consisting of 36 healthy, tumour-free testicular tissues selected from orchiectomy specimens (e.g. testicular torsions or epididymitis) were used (Table 1). As a proof of principle, a tumour group of 149 cases of primary GCT with known histological subtypes was defined as the training set (Table 1). A cohort containing 27 primary and metastatic GCTs, chosen from recent diagnostically inconclusive cases, was defined as the test set (Table 1).
CALCULATIONS AND STATISTICAL ANALYSIS
DNA quantity was calculated for each primer pair by the use of QuantStudio 5 software and individual standard curves (SCs). The SC was generated in each run by plotting the threshold cycle (Ct) values versus the dilution factor of the DNA amounts in a base-10 semilogarithmic graph. Data were applied to a straight line, and the correlation coefficient (R2) for the line was evaluated. Only SCs with a R2 of ≥ 0.99 were used. Then, the quantity for each primer pair was related to the mean quantity for all 12q primer pairs (calibrator). The resulting relative quantifications were averaged across the replicates per primer pair. Primer-specific amplification effects were estimated with linear mixed effect models (to account for repeated measures) and regressed out. The linear mixed effect models were employed by the use of r software (version 3.4.0) and the r-package lme4 (version 1.1.1319).
CGH
Array-CGH was performed on genomic DNA with the Human Genome 180 K CGH Microarray (Agilent Technologies, Waldbronn, Germany), according to the manufacturer's recommendations, as described previously.20 The arrays were scanned with the SureScan Dx Microarray scanner G5761A (Agilent Technologies, Waldbronn, Germany). Image analysis was carried out with feature extraction V12.1 (Agilent Technologies, Waldbronn, Germany), and data analysis was performed with cytogenomics V5.1.2 (Agilent Technologies, Waldbronn, Germany) (Figure S2). Interpretation was based on Human Genome Assembly 37 (GRCh37/hg19).
IMMUNOHISTOCHEMISTRY
IHC reactions were performed on 2-μm FFPE testicular tissue sections, as described previously.21 Antigen retrieval was carried out at 97°C in citrate buffer (pH 6) or EDTA buffer (pH 9). The following antibodies and dilutions were used: anti-SALL4 (monoclonal mouse, high buffer, 1:100, 30 min of incubation, clone 6E3; CellMarque, Merck KGaA, Darmstadt, Germany), anti-CDX2 (monoclonal mouse, high buffer, ready to use, 25 min of incubation, clone DAK-CDX2; Dako, Agilent Technologies, Waldbronn, Germany), anti-GPC3 (monoclonal mouse, high buffer, ready to use, 30 min of incubation, clone IGI2; DCS Innovative Diagnostik-Systeme Dr. Christian Sartori GmbH & Co. KG, Hamburg, Germany), anti-desmin (monoclonal mouse, high buffer, ready to use, 20 min of incubation, clone D33; Dako), anti-carcinoembryonic antigen (CEA) (monoclonal mouse, high buffer, ready to use, 25 min of incubation, clone II7; Dako), anti-keratin (monoclonal mouse, high buffer, ready to use, 12.5 min of incubation, clone AE1/AE3; Dako), and anti-OCT3/4 (monoclonal mouse, high buffer, ready to use, 20 min of incubation, clone N1NK; Dako). The sections were incubated with a ready-to-use horseradish peroxidase-labelled secondary antibody at room temperature for 25 min (anti-rabbit/mouse, produced in goat; REAL EnVision Detection System; Dako, Agilent Technologies, Waldbronn, Germany). The substrate DAB + Chromogen system produces a brown end product, and is applied to visualise the site of the target antigen (REAL DAB + Chromogen; Dako). Tissue samples were counterstained with Meyer's haematoxylin (Dako, Agilent Technologies, Waldbronn, Germany) for 8 min, and analysed by the use of light microscopy.
Results
ESTABLISHING THE CUT-OFF VALUE FOR i(12p) DETECTION AND EVALUATION OF THE TRAINING SET
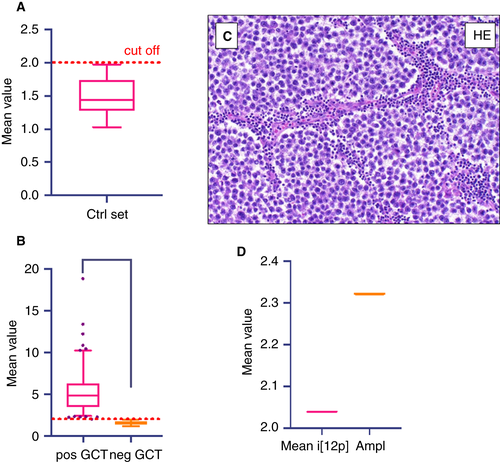
The aim of the present study was to determine the 12p DNA amount/12q DNA amount ratio. This ratio should show whether or not i(12p) is present in the analysed sample. To evaluate the 12p/12q ratio cut-off value for qPCR, we tested 36 tumour-free cases and calculated the mean value + 2 SD as the lower cut-off. The mean ratio was 1.48 and the SD was 0.257. This resulted in a cut-off amplification ratio of < 1.99 for negativity for i(12p) (Figure 2A). We then tested 149 tumour samples for i(12p), and found 133 samples (89%) to be positive and 16 samples (11%) to be negative for i(12p), as shown in Figure 2B. The mean value was 5.3, with a minimum amplification ratio of 2.02 and a maximum amplification ratio of 18.84 for the i(12p)-positive cases. All 16 tumours found to be i(12p)-negative were SEMs (Table 4).
| Case | Histology | Status | Location | i(12p) |
|---|---|---|---|---|
| 1 | Rhabdomyosarcoma (somatic malignant transformation) (Figure 3) | Primary | Testis | Yes |
| 2 | Metaplastic carcinoma (somatic malignant transformation) (Figure 4) | Primary | Testis | Yes |
| 3 | Epidermoid cyst (Figure 5) | Primary | Testis | No |
| 4 | Adenocarcinoma (somatic malignant transformation) (Figure 6) | Metastasis | Rib | Yes |
| 5 | Adenocarcinoma (somatic malignant transformation) (Figure 7) | Metastasis | Lymph node | Yes |
| 6 | Sarcomatoid yolk sac tumour (Figure 8) | Metastasis | Retroperitoneal | Yes |
| 7 | Bronchiogenic cyst (Figure 9) | Primary | Para-adrenal | No |
| 8 | Yolk sac tumour | Metastasis | Peritoneum | Yes |
| 10 | Melanoma | Metastasis | Testis | No |
| 11 | Adenocarcinoma of the prostate | Metastasis | Testis | No |
| 12 | Epidermoid cyst | Primary | Testis | No |
| 13 | Spermatocytic tumour | Primary | Testis | No |
| 14 | Epidermoid cyst | Primary | Testis | No |
| 15 | Epidermoid cyst | Primary | Testis | No |
| 16 | Yolk sac tumour | Metastasis | Lung | Yes |
| 17 | Yolk sac tumour | Metastasis | Retroperitoneal | Yes |
| 18 | Yolk sac tumour | Metastasis | Brain | Yes |
| 19 | Rhabdomyosarcoma (somatic malignant transformation) | Metastasis | Lymph node | Yes |
| 20 | High-grade sarcoma (somatic malignant transformation) | Metastasis | Lymph node | Yes |
| 21 | Adenocarcinoma (somatic malignant transformation) | Metastasis | Soft tissue | Yes |
| 22 | Adenocarcinoma (somatic malignant transformation) | Metastasis | Lymph node | Yes |
| 23 | Adenocarcinoma (somatic malignant transformation) | Metastasis | Soft tissue | Yes |
| 24 | Embryonic carcinoma | Metastasis | Lymph node | Yes |
| 25 | Yolk sac tumour | Metastasis | Lymph node | Yes |
| 26 | Yolk sac tumour | Metastasis | Lymph node | Yes |
| 27 | Yolk sac tumour | Metastasis | Lymph node | Yes |
- i(12p), isochromosome 12p.
ANALYSIS OF AMPLIFICATION OF REGION 12p12.1–12p11.2 IN i(12p)-NEGATIVE CASES
Amplification of chromosomal region 12p12.1–12p11.2 has been reported to occur in i(12p)-negative GCT cases.14, 15, 17
To investigate whether tumours without i(12p) have 12p12.1–12p11.2 amplification, we further examined the i(12p)-negative samples with an extended primer panel of five additional primer pairs corresponding to this region (Figure 1, Amp15; Table 3). With application of the established cut-off for i(12p), most of the i(12p)-negative SEMs showed no 12p12.1–12p11.2 amplification. When each case was plotted separately, one SEM showed 12p12.1–12p11.2 amplification (Figure 2C,D).
i(12p) qPCR IDENTIFIES CASES WITH UNCERTAIN GCT ORIGIN
We then performed qPCR to investigate i(12p) in 27 selected cases, comprising eight primary testicular tumours and 19 metastatic cases with ambiguous histological and IHC findings, for which a GCT origin was suspected. The results of these cases are summarised in Table 5. We found i(12p) in 19 samples, and no i(12p) in eight samples. From this sample set, we selected eight cases to further illustrate diagnostic situations in which screening for i(12p) may be warranted.
| i(12p) | Number of samples | Tumour type (n) |
|---|---|---|
| Positive | 133 | SEM (91) |
| NSE (26) | ||
| Mixed GCT (16) | ||
| Negative | 16 | SEM (16) |
- GCT, germ cell tumour; NSE, non-seminoma; SEM, seminoma.
CASES 1 AND 2
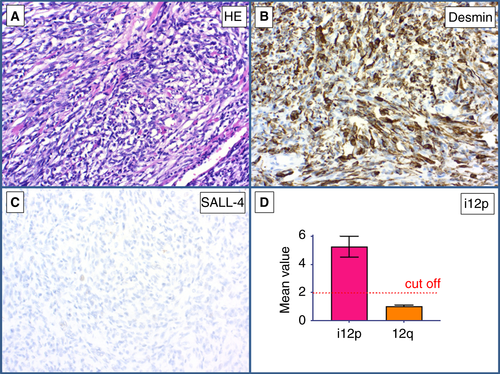
The first case was a primary testicular tumour that consisted entirely of atypical spindle cells, many of which resembled rhabdomyoblasts (Figure 3A). Immunohistochemically, the tumour cells stained positively for desmin (Figure 3B) and were negative for SALL4 (Figure 3C). qPCR showed a mean value of 5.31 for the p-arm of chromosome 12, thus proving the presence of i(12p) (Figure 3D). Therefore, this tumour was diagnosed as a rhabdomyosarcoma of GCT origin. The second case was a primary testicular tumour showing pleomorphic epithelioid morphology involving the entire testis without histological features of a GCT (Figure 4A). Immunohistochemically, the tumour showed strong positivity for pan-cytokeratin (Figure 4B) and an absence of SALL4 expression (Figure 4C). qPCR showed the presence of i(12p), with a mean value of 3.87 for the p-arm of chromosome 12, and the tumour was diagnosed as a metaplastic carcinoma of GCT origin (Table 5).
CASE 3
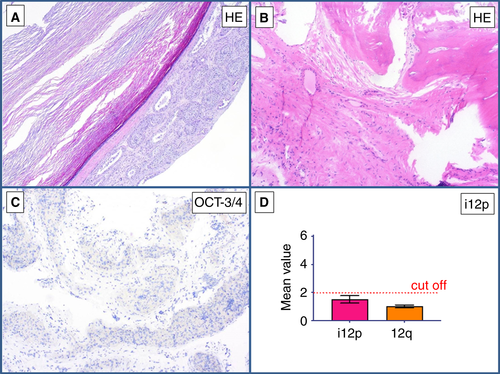
This primary tumour of the testis showed a cyst lined by squamous epithelium, partly with regressive changes and ossification (Figure 5A,B). Immunohistochemically, no surrounding GCNIS was detectable, and OCT3/4 staining was negative (Figure 5C). No i(12p) was detectable with qPCR (mean value, 1.67) (Figure 5D). On the basis of these findings, the diagnosis of an epidermoid cyst with no indication of malignancy was made (Table 5).
CASES 4 AND 5
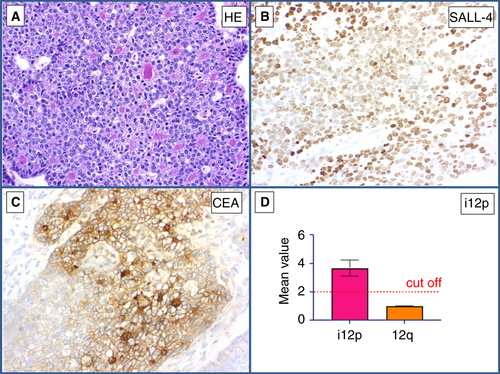
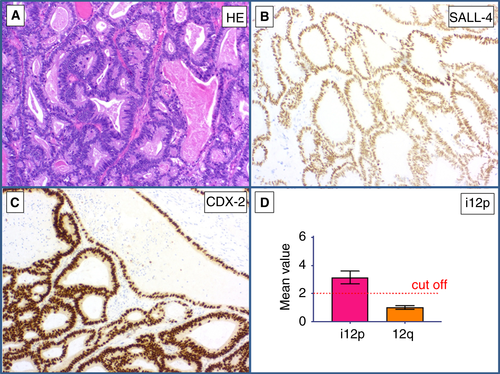
Both cases showed lymph node metastases of an adenocarcinoma (Figures 6 and 7A) in two patients with a history of GCT. Immunohistochemically, the tumour cells stained positively for SALL4 (Figures 6 and 7B), CEA (Figure 6C), and CDX2 (Figure 7C). The differential diagnosis was somatic-type malignancy after chemotherapy in a GCT versus metastases from an SALL4-positive adenocarcinoma elsewhere. qPCR showed i(12p) in both cases, with mean values of 3.24 (Figure 6D) and 3.51 (Figure 7D), respectively. Hence, the diagnosis of a somatic malignant transformation (GCT to adenocarcinoma) was made (Table 5).
CASE 6
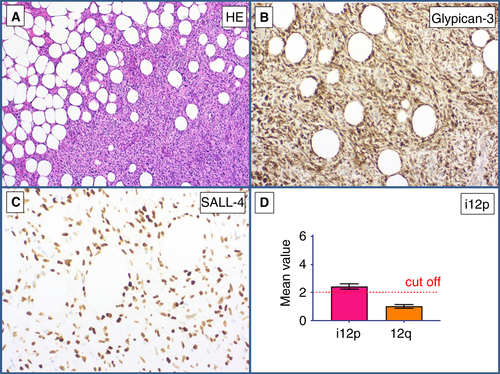
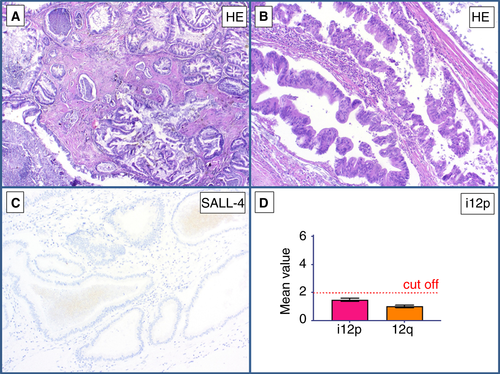
A metastasis of a sarcomatoid tumour (Figure 8A) that infiltrated the adjacent adipose tissue was found in a retroperitoneal lymph node in a patient with a history of GCT. The differential diagnosis was metastatic GCT with somatic-type malignancy versus primary retroperitoneal soft tissue sarcoma. Immunohistochemically, the tumour cells were positive for SALL4 (Figure 8B) and GPC3 (Figure 8C). MDM2 FISH analysis gave a negative result, thus ruling out a dedifferentiated liposarcoma. qPCR showed the presence of i(12p) (mean value of 2.34 for the p-arm of chromosome 12) (Figure 8D). On the basis of this finding, the diagnosis of a sarcomatoid YST was made (Table 5).
CASE 7
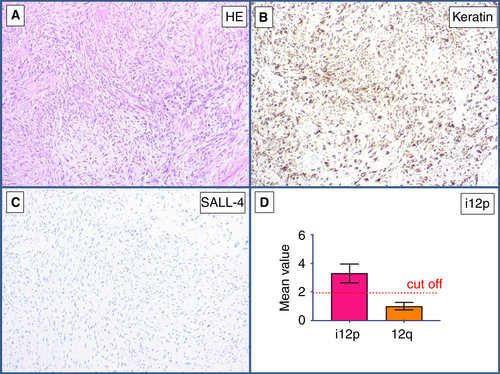
This intra-abdominal tumour showed mature epithelial structures with neither cellular nor nuclear atypia (Figure 9A). The epithelial structures showed ciliated epithelium (Figure 9B). No SALL4 positivity was detected immunohistochemically (Figure 9C). qPCR showed a mean value of 1.77 for the p-arm of chromosome 12, and thus an absence of i(12p) (Figure 9D). In this case, the diagnosis of an intra-abdominal bronchogenic cyst was made (Table 5).
Discussion
Since its first description in malignant type II GCTs> 35 years ago,10 i(12p) has been the subject of substantial investigation. Today, it seems certain that i(12p) plays a decisive role in the transition from GCNIS to invasive GCT.22 Different methods have been used for the detection of i(12p), such as FISH,11 CGH,13 and PCR.17 So far, qPCR has had no diagnostic value in the molecular analysis of i(12p) in GCT. To date, FISH has remained the predominant method for screening for chromosomal aberrations within GCTs.7, 23, 24
In this study, we used qPCR as a new method for detecting i(12p) in a large cohort of primary GCTs. We found that i(12p) was present in the majority of GCTs (Table 4). Thus, the presence of i(12p) is a valuable marker for GCT in diagnostic surgical pathology. Most GCTs are readily diagnosed on the basis of histological features and IHC staining. Nevertheless, there are cases in which conventional methods cannot prove or rule out a GCT origin. Cases with somatic malignant transformation or metastases are particularly difficult to diagnose, as has also been shown here.25, 26 Worth mentioning are adenocarcinomas or their metastases, especially in retroperitoneal or mediastinal locations, with stem cell-like or fetal-like features, which can be challenging to differentiate from GCTs with somatic malignant transformation. The reason is their possible coexpression of the stem cell factor SALL4 and other germ cell markers, i.e. α-fetoprotein and GCP3.27, 28 Therefore, qPCR, as an additional detection method, is a useful tool to aid in the diagnosis. Demonstration of the presence of i(12p) as a strong indicator of a GCT22 was an important prerequisite for diagnosing the presented exemplary clinical cases.
COMPARISON OF qPCR AND FISH IN i(12p) DETECTION
qPCR has been a reliable and safe method in scientific and diagnostic investigations for many years.29, 30 Its value lies primarily in the direct measurement of the multiplied chromosome sections and the direct interpretation of these results.31 Thus, qPCR seems to be particularly suitable for screening for highly frequent chromosomal aberrations, such as i(12p) in GCT. Furthermore, its implementation in routine practice is quite simple, time-saving, and inexpensive. In addition, DNA extraction and further qPCR examinations are possible on very small tissue samples, and this can be important for punch biopsies (on which we have not yet focused). In our study, the average DNA amount of tissue samples was 82.75 ng/µl (lowest, 8.8 ng/µl; highest, 361 ng/µl; data not shown), but small amounts of DNA were also successfully analysable, and the age of the paraffin-embedded tissue did not matter. FISH analysis has now been used for several years, and is considered to be the gold standard. The advantage of FISH is that the chromosomal change in a GCT sample can be detected directly with a specific probe.32 However, FISH requires the correct assessment of a representative number of tumour cells by a trained expert,17, 24 whereas qPCR can be performed by technical staff without specific training on tissue sections. In the current study, we have shown that the rate of detection of i(12p) with qPCR is as great as that obtained with FISH.23 Therefore, we propose qPCR as an equivalent assay for the detection of i(12p) in extragonadal manifestations of GCT (e.g. metastases and primary extragonadal GCT). Furthermore, FISH analysis is not available in all pathological institutions.33 The specificity of the qPCR method is defined by the specificity of the four PCR products for i(12p). Melting curve analyses resolved a single amplicon for each primer pair. In addition, the differences in the amplification value for the four amplicons showed a comparable range and a similar pattern over the samples. To exclude false-negative cases and to confirm the results of qPCR, we performed array-CGH analysis as described above. Two cases of tumour-free testicular tissue, two non-GCTs and two GCTs without i(12p) on qPCR analysis did not show 12p gain in array-CGH analysis (Figure S2). In contrast, two GCT cases (NSE and SEM) showed 12p gain in array-CGH analysis (Figure S2). Thus, the results of array-CGH confirmed the results of qPCR.
The data obtained in this study also allowed us to determine the prevalence of i(12p). Our results indicate that the previously reported prevalence of i(12p) in 80% of type II GCTs7, 34 appears to be too low. Instead, we found a prevalence of 89%, which is in close accordance with a FISH-based publication from Shen et al., showing i(12p) in 87% of GCTs.23
i(12p)-NEGATIVE CASES
In a few cases without i(12p), the diagnosis remained problematic. Our extended qPCR method investigating the 12p11.2–12.1 region was not helpful in these cases. MicroRNAs have been shown to be more accurate, as these markers are expressed in all GCTs regardless of their i(12p) status.23 miR-371a-3p and miR-375 have high specificity and sensitivity for type II GCT.23, 35 MiR-371a-3p is not expressed in TEs,36 and is therefore presumably also not expressed in somatic malignant transformation (SMT) arising from TE components. Thus, both miRNA markers must be investigated in tumour tissue from TEs or cases with SMT.
In addition to the molecular genetic studies of overrepresented genetic areas, we carried out a histological and IHC analysis of all i(12p)-negative cases. Lymphocytic infiltrates with a granulomatous inflammation pattern were found in all tissue samples. These observations were also made in other studies.23, 37 The body's immune response can lead to tumour regression within the testis. Therefore, some GCTs only become clinically apparent after they have metastasised.25, 38 Whether the immunological response to tumour tissue may be related to the lack of i(12p) has not been investigated, and merits further study. However, lymphocytic infiltrates have been described in both i(12p)-negative and i(12p)-positive cases. A causal relationship between lymphocytic infiltrates and the lack of i(12p) cannot be established on the basis of the observations in this work. Nevertheless, it is striking that Shen et al. made the same observations regarding the prevalence of i(12p)-positive versus i(12p)-negative cases, and the occurrence of lymphocytic infiltrates in GCT.23 It should be particularly emphasised that the i(12p)-negative cases in the studies by Shen et al. were exclusively SEMs.39 These consistent observations in two independent molecular genetic studies are remarkable, and justify further research on i(12p)-negative GCTs.
i(12p) AND GCNIS
In the past, only a few GCNIS cases with and without adjacent invasive GCT were investigated, and they had ambiguous results for the presence of i(12p) or the amplification of 12p material in GCNIS cells. However, it is well known that these genomic changes play an important role in the development of invasion.8, 22, 40 As we have only investigated one GCNIS case without adjacent invasive GCT, which was i(12p)-negative, we cannot give much information about the molecular characteristics of GCNIS. This subject should be investigated in further studies.
CHALLENGING CASES AND CLINICAL RELEVANCE OF THE CORRECT DIAGNOSIS
i(12p) specifies a GCT-associated origin, whereas somatic tumours (e.g. colorectal or lung cancer) do not show gain of 12p material, as shown by Kernek et al.41 Despite an ongoing debate, GCT patients with somatic malignant transformation may benefit from GCT-specific cisplatin-based chemotherapy approaches, as histology-driven treatment regimens, as used for cancer of unknown primary, do not consistently translate into improved outcomes.42-44 Therefore, it is of great importance for the determination of treatment strategy to confirm a GCT origin and rule out a somatic primary, if patients with a history of GCT develop solitary nodules suspicious of tumour in the retroperitoneum, the lung or the mediastinum with non-germ cell cancer histology. Additionally, surgical salvage approaches in cases of limited metastatic disease at relapse seem to be justified if complete resection appears feasible, which is in accordance with late relapses of classic GCTs. Moreover, there is an increased risk of developing further i(12p)-related malignancies, including haematological neoplasia, such as acute myeloid leukaemia, particularly in patients with primary mediastinal GCTs, which confers a substantially poorer prognosis.45, 46
In the test cohort, 16 metastases of GCT (Table 4), seven of them as somatic malignant transformation, could be confirmed. All of them developed after chemotherapy and contained i(12p). This suggests persistence of i(12p) in the metastatic process without an influence of chemotherapy. In their series of sarcomatoid YSTs, Idrees et al. had one patient with two i(12p)-positive metastases, and one case in which the first and the fourth YST recurrences were i(12p)-negative, and the third and fifth recurrences were i(12p)-positive.24 However, in our investigation, we did not find i(12p) loss in the tissue specimens of GCT after chemotherapy.
In summary, this study shows that qPCR is a simple and cost-effective method for the detection of i(12p), as a useful addition to the standard pathological evaluation, and greatly assists in the diagnosis of challenging tumours with a possible GCT origin. We detected i(12p) in 89% of the GCT cases, suggesting that the i(12p) frequency is higher than previously reported. All i(12p)-negative cases were SEMs.
Acknowledgements
F. Bremmer is supported by the Wilhelm Sander-Stiftung (Grant numbers 2016.041.1 and 2016.041.2). We thank Olga Dschun for excellent technical assistance. F. Bremmer, D. Nettersheim and C. Oing took part in this study as members of the German Testicular Cancer Study Group (GTCSG) of the Deutsche Krebsgesellschaft. Open access funding enabled and organized by ProjektDEAL.
Conflicts of interest
The authors declare no conflicts of interest.
Author contributions
F. Bremmer conceived and designed the study, collected and analysed the data, performed histological analyses, and wrote the manuscript. A. Fichtner, A. Richter and S. Kaulfuß conceived and designed the study, collected and analysed the data, and wrote the manuscript. S. Filmar collected and analysed the data, and performed statistical analyses. N. T. Gaisa, S. Schweyer and H. Reis performed histological analysis, and edited the manuscript. D. Nettersheim and C. Oing interpreted the data, and performed critical revision for important intellectual content. F. A. Gayer reviewed clinical records. A. Leha performed statistical analyses. P. Ströbel performed critical revision for important intellectual content. All authors gave final approval for publication.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.




