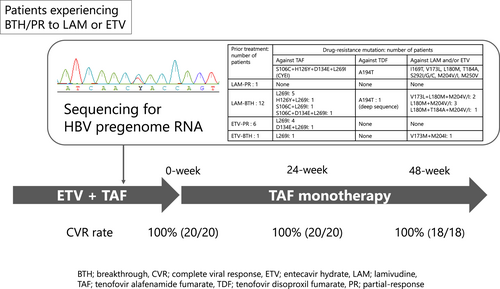
Switching from combination therapy with entecavir hydrate plus tenofovir alafenamide fumarate to tenofovir alafenamide fumarate monotherapy in patients with chronic hepatitis B based on nucleotide sequences of hepatitis B virus pregenome RNA
Abstract
Aim
Patients with chronic hepatitis B virus (HBV) infection experiencing viral breakthrough (BTH) or partial response (PR) during lamivudine (LAM) or entecavir hydrate (ETV) administration often took ETV plus tenofovir alafenamide fumarate (TAF) due to the emergence of a drug-resistance mutation. However, in patients lacking drug-resistance mutation against TAF, sufficient antiviral effects may be achievable with TAF monotherapy. We assessed the drug-resistance profile through nucleotide sequences of HBV pregenome RNA, and subsequently changed to TAF monotherapy from ETV plus TAF.
Methods
This prospective study included 25 patients with serum HBV-DNA below 20 IU/mL under ETV plus TAF administration. Pregenome RNA nucleotide sequences of HBV in the sera were analyzed using direct sequencing and deep sequencing. ETV was discontinued in patients without rtA194T and rtS106C + rtH126Y + rtD134E + rtL269I quadruple mutations in direct sequencing.
Results
LAM-PR, LAM-BTH, ETV-PR, and ETV-BTH were observed in 1, 16, 7, and 1 patient(s), respectively. Pregenome RNA nucleotide sequences were analyzable in 20 patients. Among the 12 patients classified as LAM-BTH, six patients showed rtL180M + rtM204V/I in direct sequencing, and one patient showed minor clones containing rtL180M + rtM204V + A194T in deep sequencing at a frequency of 0.3%. In the six patients classified as ETV-PR, one patient harbored rtM204I. No clones showing rtS106C + rtH126Y + rtD134E + rtL269I quadruple mutation were detected in deep sequencing. Subsequently, ETV was discontinued, and serum HBV-DNA remained undetectable up to 48 weeks in all patients.
Conclusion
Patients receiving ETV plus TAF due to partial response or BTH during initial LAM or ETV administration were able to safely transition to TAF monotherapy based on nucleotide sequences of HBV pregenome RNA in the sera.
Graphical Abstract
We devised a method to analyze drug resistance profile of hepatitis B virus pregenome RNA even when serum hepatitis B virus DNA is undetectable during antiviral therapy. By using this approach, we verified that patients receiving combination therapy with entecavir hydrate plus tenofovir alafenamide fumarate for chronic hepatitis B virus infection due to experiencing viral breakthrough or partial response during initial lamivudine or entecavir hydrate administration were able to safely transition to tenofovir alafenamide fumarate monotherapy.
Abbreviations
-
- ADV
-
- adefovir pivoxil
-
- ALT
-
- alanine aminotransferase
-
- BTH
-
- breakthrough
-
- CKD
-
- chronic kidney disease
-
- CVR
-
- complete viral response
-
- CYEI
-
- rtS106C + rtH126Y + rtD134E + rtL269I
-
- eGFR
-
- estimated glomerular filtration rate
-
- ETV
-
- entecavir hydrate
-
- HBcrAg
-
- hepatitis B core-related antigen
-
- HBeAg
-
- hepatitis B e antigen
-
- HBsAg
-
- hepatitis B surface antigen
-
- HBV
-
- hepatitis B virus
-
- HCC
-
- hepatocellular carcinoma
-
- HCV
-
- hepatitis C virus
-
- HIV
-
- human immunodeficiency virus
-
- jRCT
-
- the Japan Registry of Clinical Trials
-
- LAM
-
- lamivudine
-
- NA
-
- nucleos(t)ide analog
-
- Pol/RT
-
- polymerase/reverse transcriptase
-
- PR
-
- partial response
-
- TAF
-
- tenofovir alafenamide fumarate
-
- TDF
-
- tenofovir disoproxil fumarate
-
- TFV
-
- tenofovir
INTRODUCTION
In antiviral therapy for patients infected with hepatitis B virus (HBV), administration of nucleos(t)ide analogs (NAs) exert potent antiviral effects regardless of HBV genotypes and host factors. NAs inhibit the reverse transcription of HBV-DNA within hepatocytes, thereby suppressing the release of HBV-DNA into the bloodstream and leading to the resolution of hepatitis, as well as reducing the progression of liver fibrosis and development of hepatocellular carcinoma (HCC). However, NAs cannot eliminate covalently closed circular DNA from hepatocytes. As a result, patients with chronic hepatitis B require lifelong administration of NAs to prevent the risk for reactivation and/or severe exacerbation of hepatitis on discontinuation of treatment.1, 2
Long-term administration of lamivudine (LAM) has been reported to lead to the emergence of drug-resistant mutations, such as rtL180M and rtM204V/I.3, 4 Several studies have demonstrated that entecavir hydrate (ETV) shows mild cross-resistance to LAM-resistant mutations, such as rtL180M and rtM204V/I,5, 6 whereas tenofovir (TFV) does not.7 However, it has been reported that the rtA194T mutation reduces TFV sensitivity by increasing the IC50 level in in vitro analyses.8, 9 A recent study reported that the combination of quadruple mutations, namely rtS106C, rtH126Y, rtD134E, and rtL269I (CYEI) mutation, leads to a 15.3-fold increase in the amount of TFV required to inhibit HBV at the IC50 level, and a 26.3-fold increase at the IC90 level in in vitro analyses.10 Thus, in patients currently receiving combination therapy with ETV and tenofovir alafenamide fumarate (TAF) after the initial LAM administration, if LAM-resistant mutations were to emerge, it is possible that antiviral efficacy would still be favorable even with TAF monotherapy, provided that there exist no HBV clones harboring the rtA194T or CYEI mutations.
In clinical practice, analysis for drug-resistant mutations in HBV genome is challenging due to the difficulty of genome sequencing of HBV-DNA when there is a good antiviral response with undetectable serum HBV-DNA. Thus, we focused on HBV-RNA. Recently, serum HBV-RNA has been proposed as a novel surrogate biomarker of covalently closed circular DNA levels in hepatocytes, and serum HBV-RNA has been shown to be useful in various clinical contexts, such as the monitoring transcriptional activity of HBV and the discontinuation of NAs during long-term anti-viral therapy.11-13 We devised a method to sequence the HBV pregenome RNA secreted in the sera during NAs administration. This approach enables the analysis of HBV genome sequences even when serum HBV-DNA is undetectable during antiviral therapy with NAs. Thus, the aim of the present study was to analyze the antiviral efficacy after switching to TAF monotherapy from the combination therapy of TAF and ETV in patients with chronic hepatitis B who initially had a poor antiviral response after LAM or ETV administration, but currently show a good antiviral response while receiving combination therapy with ETV and TAF, provided that no drug resistance mutations against TAF and tenofovir disoproxil fumarate (TDF) were detected by sequencing HBV pregenome RNA in the sera.
METHODS
Patients and study design
This was a single-arm prospective interventional study conducted at Saitama Medical University Hospital, Iruma-gun, Japan. The study enrolled patients with chronic HBV infection who were receiving combination therapy with ETV plus TAF and showing complete viral response defined as serum HBV-DNA level <20 IU/mL. The enrollment period was between March 2022 and June 2022. Patients with coinfection of either hepatitis C virus or HIV, those receiving immunosuppressive agents, and those with underlying decompensated cirrhosis and/or end-stage HCC were excluded from the study.
First, amino acid sequences of the polymerase/reverse transcriptase (Pol/RT) domain in HBV pregenome RNA were evaluated using direct-sequencing and deep-sequencing methods. Second, in patients without TAF-resistant or TDF-resistant mutations in the Pol/RT domain, the NAs used for the treatment were switched from ETV plus TAF combination therapy to TAF monotherapy. Patients who developed viremia with HBV-DNA levels exceeding 2000 IU/mL after switching to TAF monotherapy resumed combination therapy with ETV and TAF.
The primary endpoint was complete viral response rate at 4, 12, 24, and 48 weeks after switching to TAF monotherapy. The secondary endpoints were alanine aminotransferase (ALT) levels at 4, 12, 24, and 48 weeks, and change in renal function, as measured by estimated glomerular filtration rate (eGFR), and serum phosphorus levels. In addition, the changes in eGFR and serum phosphorus levels were compared for 48 weeks before and after switching to TAF monotherapy.
Evaluation of the virologic markers for HBV
Serum HBV-DNA levels were measured using the COBAS TaqMan HBV Test, v2.0 (Roche Diagnostics K.K.), with a lower limit of quantification of 20 IU/mL. Serum hepatitis B surface antigen (HBsAg) levels were measured using the Architect HBsAg QT assay kit (Abbott Japan Corp.) through chemiluminescent immunoassay. Hepatitis B e antigen (HBeAg), anti-HBe, and hepatitis B core-related antigen (HBcrAg) were measured using chemiluminescence enzyme immunoassay with the Architect HBeAg (Abbott Japan Corp.), Architect HBeAb (Abbott Japan Corp.), and iTACT HBcrAg (Fuji-Rebio) kits,14 respectively. The HBV genotypes were identified using enzyme-linked immunosorbent assay with monoclonal antibodies targeting type-specific epitopes in the preS2-region (Institute of Immunology).
Sequencing of Pol/RT domain in HBV pregenome RNA
To analyze the amino acid sequences of the Pol/RT domain, HBV viral DNA/RNA containing the pregenome RNA was extracted from the serum at a volume of 200 μL using the QIAamp MinElute Virus Spin Kit (Qiagen K.K.), and DNA was digested using Recombinant DNase I (TaKaRa Bio Inc.). Reverse transcription-polymerase chain reaction (RT–PCR) and nested PCR were performed with the following primer sets and conditions. In the RT–PCR, the primer set of BYs-1L/BYs-1R was used with PrimeScript II High Fidelity One Step RT-PCR Kit (TaKaRa Bio Inc.). The reverse transcription step involved a reaction at 45°C for 10 min, followed by a denaturation at 94°C for 2 min. The subsequent PCR steps involved a denaturation at 98°C for 10 s, an annealing at 60°C for 15 s, and an extension at 68°C for 10 s. This PCR step was repeated 40 times. In the nested PCR, the primer set of BYs-NGS-L/BYs-NGS-R was used with PrimeSTAR Max DNA Polymerase (TaKaRa Bio Inc.). The subsequent PCR steps involved a denaturation at 98°C for 10 s, an annealing at 55°C for 5 s, and an extension at 72°C for 5 s. This nested PCR step was repeated 30 times. Detailed primer sequences and amino/nucleotide positions are shown in Table 1.
| Name | Nucleoside sequences (5′→3′) | aa position in RTa | nt position in HBVa |
|---|---|---|---|
| BYs-1L | ATCGYTGGATGTGTCTGCGGCGTTT | rt80-rt88 | nt369-nt393 |
| BYs-1R | AAGGCCTTGTAAGTTGGCGAGAAAG | rt322-rt330 | nt1094-nt1118 |
| BYs_NGS-L | TATGCCTCATCTTCTTGTTGGTTCT | rt97-rt105 | nt420-nt444 |
| BYs_NGS-R | ATTKACAGGCAGTTTYCGAAAACAT | rt271-rt279 | nt942-nt966 |
- Abbreviations: aa, amino acids; HBV, hepatitis B virus; nt, nucleotides; RT, reverse transcription.
- a Nucleotide position 1 is designated as the EcoRI restriction enzyme site.
Direct sequencing was performed using the sequencing primers of BYs-NGS-L and BYs-NGS-R, the same ones used in the nested PCR on a 3130 Genetic Analyzer (Thermo Fisher Scientific K.K.). The resulting nucleotide sequences were assembled using ATGC for Windows (GENETYX Corp.). Amplicon deep sequencing was conducted using MiSeq (Illumina). The cut-off value was set at 0.2% relative to the total HBV pregenome RNA. In brief, a fragment with a length of 215 amino acids corresponding to the sequence between rt106 and rt270 was analyzed. All work processes were performed according to the manufacturer's protocol.
The amino acid sequences of major clones and drug resistance clones were assessed by referring to the sequences of GenBank accession number from LC784158 to LC784243 and LC787270, LC787271.
Definition of drug-resistant mutation in the Pol/RT domain
Drug-resistant mutations in the Pol/RT domain, such as aa106, aa126, aa134, aa169, aa173, aa180, aa181, aa184, aa194, aa202, aa204, aa236, aa250, and aa269, were evaluated. CYEI mutation was defined as TAF-resistance mutation, rtA194T was defined as TDF-resistance mutation, and I169T, V173L, L180M, A181T/V, T184S/G/C/A/I/L/M/F, S202I/G/C, M204V/I, and M250V were defined as LAM-resistance and/or ETV-resistance mutations.
Statistical analysis
Categorical data were compared using Fisher's exact test. The distributions of continuous variables were analyzed using the Mann–Whitney's U-test. Differences in continuous variables were assessed using Wilcoxon's signed-rank test. All tests of significance were two-tailed, and p-values <0.05 were considered statistically significant. JMP® Pro software version 16.0.0 (SAS Institute Inc.) was utilized for the analyses.
RESULT
Demographic characteristics and clinical features of patients
A total of 25 patients were enrolled in the study, consisting of 13 men (52.0%) and 12 women (48.0%). The median age of the patients was 61 years (range 41–88 years). The demographic characteristics and clinical features of the patients at baseline are presented in Table 2. The HBV genotype distribution among the patients was as follows: three patients (12.0%) had genotype B, 21 patients (84.0%) had genotype C, and the genotype was undetermined in one patient (4.0%). Regarding prior treatment histories, 17 patients (68.0%) had received LAM as the first-line drug, with one patient showing a partial response (LAM-PR) and 16 patients experiencing viral breakthrough (LAM-BTH). Eight patients (32.0%) had received ETV, out of which seven patients showed a partial response (ETV-PR), and one patient had viral breakthrough (ETV-BTH). All 25 patients had received a combination of ETV and TAF, and had achieved HBV-DNA levels below 20 IU/mL. The median duration for which serum HBV-DNA levels remained below the detection sensitivity was 149.3 months (range 57.5–227.1 months). Serum HBeAg testing was positive in 12 patients (48.0%). The median HBsAg level was 1143.78 IU/mL (range 0.04–5827.06 IU/mL), and the median serum HBcrAg level was 4.2 Log U/L (range <2.1–6.9). None of the patients had underlying compensated cirrhosis or received previous therapy for HCC.
| Factors | Overall (n = 25) | Patients with unsuccessful RNA amplification (n = 5) | Patients with successful RNA amplification (n = 20) | p-value |
|---|---|---|---|---|
| Age (years) | 61 (41–88) | 68 (49–88) | 59 (41–79) | 0.0663 |
| Male : female | 13 (52%):12 (48%) | 4 (80%):1 (20%) | 9 (45%):11 (55%) | 0.1485 |
| Genotype B : C: undetermined | 3 (12%):21 (84%):1 (4%) | 2 (40%):2 (40%):1 (20%) | 1 (5%):19 (95%):0 (0%) | 0.0085 |
| HBeAg-positive | 12 (48%) | 0 (0%) | 12 (60%) | 0.0055 |
| HBsAg (IU/mL) | 1143.78 (0.04–5827.06) | 1.69 (0.04–1299.17) | 1549.86 (11.35–5827.06) | 0.0080 |
| iTACT-HBcrAg (U/L) | 4.2 (<2.1–6.9) | 3.0 (<2.1–4.2) | 5.2 (2.6–6.9) | 0.0143 |
| LAM-PR : LAM-BTH : ETV-PR : ETV-BTH | 1 (4%):16 (64%):7 (24%):1 (8%) | 0 (0%):4 (80%):0 (0%):1 (20%) | 1 (5%):12 (60%):6 (30%):1 (5%) | 0.3728 |
| Patients achieving HBV-DNA levels <20 IU/mL | 25 (100%) | 6 (100%) | 19 (100%) | 1.0000 |
| Period achieving HBV-DNA levels <20 IU/mL (months) | 149.3 (57.5–227.1) | 151.9 (120.3–214.7) | 147.9 (57.5–227.1) | 0.2481 |
| Platelet count (104/mm3) | 22.4 (13.9–45.2) | 20.7 (15.1–30.4) | 22.7 (13.9–45.2) | 0.6818 |
| Albumin (g/dL) | 4.3 (3.7–5.0) | 4.1 (3.9–4.7) | 4.3 (3.7–5.0) | 0.5493 |
| AST (U/L) | 22 (13–113) | 24 (17–27) | 21 (13–113) | 0.4316 |
| ALT (U/L) | 16 (10–73) | 14 (12–21) | 17 (10–73) | 0.3498 |
| Creatinine (mg/dL) | 0.79 (0.54–1.50) | 0.76 (0.62–1.50) | 0.79 (0.54–1.40) | 0.7092 |
| eGFR (mL/min/1.73 m2) | 70.1 (34.3–103.8) | 73.3 (34.3–85.2) | 68.8 (40.0–103.8) | 1.0000 |
| Total cholesterol (mg/dL) | 186 (126–258) | 178 (171–219) | 186 (126–258) | 0.9405 |
| Triglyceride (mg/dL) | 106 (52–181) | 92 (52–118) | 108 (60–182) | 0.1359 |
| Alpha fetoprotein (ng/mL) | 2.5 (2.0–6.7) | 2.7 (2.0–6.7) | 2.4 (2.0–5.4) | 0.4692 |
- Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; BTH, breakthrough; eGFR, estimated glomerular filtration rate; ETV, entecavir hydrate; HBcrAg, hepatitis B core-related antigen; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; LAM, lamivudine; PR, partial response.
Factors associated with successful amplification of HBV pregenome RNA
Amplification of HBV pregenome RNA was successfully achieved in 20 patients (80.0%) using RT–PCR and second PCR. The frequency of patients with genotype C and positive HBeAg was higher in those with successful RNA amplification compared with the remaining five patients (95.0% vs. 40.0%, p = 0.0085 and 60.0% vs. 0%, p = 0.0055). Additionally, the levels of HBsAg and HBcrAg were higher in the 20 patients with successful RNA amplification compared with the remaining five patients (Table 2). There was no association observed between achievement of pregenome RNA amplification and laboratory values other than viral markers.
Direct sequencing of Pol/RT domain in HBV pregenome RNA
Among the 20 patients in whom amplification of HBV pregenome RNA was achieved, direct sequencing revealed the presence of rtS106C in two patients (10.0%), rtH126Y in one patient (5.0%), rtD134E in two patients (10.0%), and rtL269I in 14 patients (70.0%). As for the combination of these four mutations, one patient (8.3%) showed triple mutations of rtS106C + rtD134E + rtL269I, and each one patient (8.3%) showed double mutations of rtS106C + rtL269I and rtH126Y + rtL269I in the LAM-BTH group. Additionally, one patient (16.7%) in the ETV-PR group showed double mutations of rtD134E + rtL269L. In contrast, no patient showed quadruple mutations of rtS106C + rtH126Y + rtD134E + rtL269I (Table 3).
| Prior treatment: No. patients | Drug-resistance mutation: No. patients | ||
|---|---|---|---|
| Against TAF | Against TDF | Against LAM and/or ETV | |
| S106C + H126Y + D134E + L269I (CYEI) | A194T | I169T, V173L, L180M, T184A, S292I/G/C, M204V/I, M250V | |
| LAM-PR: 1 | None | None | None |
| LAM-BTH: 12 | L269I: 5 | None | V173L + L180M + M204V/I: 2 |
| H126Y + L269I: 1 | |||
| S106C + L269I: 1 | L180M + M204V/I: 3 | ||
| S106C + D134E + L269I: 1 | L180M + T184A + M204V/I: 1 | ||
| ETV-PR: 6 | L269I: 4 | None | None |
| D134E + L269I: 1 | |||
| ETV-BTH: 1 | L269I: 1 | None | V173M + M204I: 1 |
- Abbreviations: BTH, breakthrough; ETV, entecavir hydrate; LAM, lamivudine; PR, partial response; TAF, tenofovir alafenamide fumarate; TDF, tenofovir disoproxil fumarate.
In contrast, with regard to resistance mutations against LAM and/or ETV, six patients (50.0%) in the LAM-BTH group showed both rtL180M and rtM204V/I, two (16.7%) of whom also had rtV173L, and one patient (8.3%) had rtT184A. Furthermore, one patient (100%) in the ETV-BTH group showed rtV173L in addition to rtM204I (Table 3).
No significant associations were observed between the presence of rtS106C and/or rtH126Y and/or rtD134E and/or rtL269I and the presence of rtL180M and/or rtM204V/I.
Deep sequencing of Pol/RT domain in HBV pregenome RNA
In deep sequencing, amplicon data with an average read depth of 232 413 were obtained from 20 patients (Table 4). Drug-resistance mutations detected by direct sequencing were also identified. However, in addition to these mutations, a minor clone harboring rtH126Y with a relative frequency of 0.40% of the total pregenome RNA was found in one patient in the LAM-BTH group. Similarly, another patient in the ETV-PR group (BY-22) had a minor clone harboring rtS106C at a frequency of 0.77%. No patients were found to have clones harboring all four mutations of rtS106C + rtH126Y + rtD134E + rtL269I, even with deep sequencing.
| Prior treatment histories | Total read | Drug-resistance mutation | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S106C | H126Y | D134E | L269I | L180M | M204V/I | V173L/M | T184A | A194T | ||
| LAM-PR | 261 553 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| LAM-BTH | 209 222 | 0.0% | 0.0% | 0.0% | 63.2% | 69.7% | 71.8% | 27.4% | 0.0% | 0.0% |
| 258 475 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| 293 901 | 0.0% | 0.0% | 100.0% | 100.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| 223 942 | 0.0% | 100.0% | 0.0% | 47.7% | 47.6% | 47.6% | 0.0% | 47.6% | 0.0% | |
| 198 595 | 9.1% | 0.0% | 31.4% | 3.9% | 37.3% | 51.0% | 0.0% | 0.0% | 0.0% | |
| 258 860 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.3% | 0.0% | 0.0% | |
| 174 191 | 0.0% | 0.0% | 0.0% | 3.3% | 51.6% | 93.2% | 2.4% | 0.0% | 0.0% | |
| 170 033 | 0.0% | 0.4% | 0.0% | 74.3% | 79.3% | 79.5% | 79.6% | 0.4% | 0.3% | |
| 248 487 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| 226 914 | 100.0% | 0.0% | 0.0% | 100.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| 231 062 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.2% | 0.0% | 0.0% | |
| 265 567 | 0.0% | 0.0% | 100.0% | 100.0% | 100.0% | 100.0% | 0.0% | 0.0% | 0.0% | |
| ETV-PR | 222 332 | 0.0% | 0.0% | 20.2% | 9.7% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| 214 243 | 0.0% | 0.0% | 0.0% | 100.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| 162 483 | 0.0% | 0.0% | 0.0% | 100.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| 302 428 | 0.0% | 0.0% | 0.0% | 100.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| 251 752 | 0.8% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| 213 558 | 0.0% | 0.0% | 0.0% | 100.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| ETV-BTH | 260 680 | 0.0% | 0.0% | 0.0% | 100.0% | 0.0% | 100.0% | 12.2% | 0.0% | 0.0% |
- Abbreviations: BTH, breakthrough; ETV, entecavir hydrate; LAM, lamivudine; PR, partial response.
In the LAM-BTH group, three patients had minor clones harboring V173L/M with frequencies of 0.25%, 0.26%, and 2.40%, respectively. Interestingly, in one patient from the LAM-BTH group, a minor clone harboring T184A (LC787270) at a frequency of 0.36% and A194T (LC787271) at a frequency of 0.30% was identified. Amplicon sequencing analysis revealed that rtT184A and rtA194T existed in different clones in this patient. Furthermore, rtA194T coexisted with rtrV173L, rtL180M, and rtM204V in the same clone, at a frequency of 0.30%. However, rtS106C, rtH126Y, rtD134E, and rtL269I were not found in this clone.
Viral response and ALT level in patients who switched from combination therapy with ETV and TAF to TAF monotherapy
TAF monotherapy was carried out in 20 patients whose pregenome RNA was sequenced in the sera. Two patients were excluded during follow-up: one due to development of HCC, and the other due to the initiation of pegylated interferon-α-2a therapy. Consequently, 18 patients were available for the analysis.
After the initiation of TAF monotherapy, all patients successfully maintained complete viral response for 48 weeks without viremia relapse (Figure 1a). Notably, even in the patient in whom rtA194T was detected at a frequency of 0.3% by deep sequencing. Furthermore, an additional analysis of HBV-RNA sequencing was conducted at 48 weeks after initiating TAF monotherapy, enabling HBV-RNA sequencing in a total of 10 patients. Among these, some nucleotide substitutions were observed in two patients harboring L180M + M204V mutations. However, no new amino acid substitutions were identified in any of the patients after initiating TAF monotherapy.
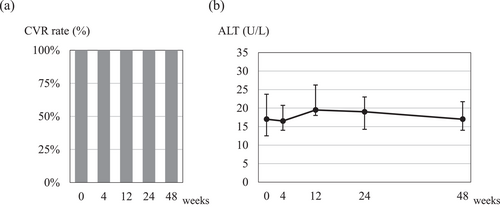
Viral response and alanine aminotransferase (ALT) levels in patients who switched from combination therapy with entecavir hydrate plus tenofovir alafenamide fumarate to tenofovir alafenamide fumarate monotherapy. (a) Complete viral response rate, (b) ALT levels at 4, 12, 24, and 48 weeks after switching to tenofovir alafenamide fumarate monotherapy. The points and error bars represent the median values and quartiles (Q1 and Q3).
The median serum ALT levels (U/L) at 0, 4, 12, 24, and 48 weeks were 17 (IQR 13–24), 17 (14–21), 20 (18–26), 19 (14–23), and 17 (14–22), respectively. These levels remained consistent until 48 weeks after initiating TAF monotherapy (Figure 1b). Importantly, none of the patients experienced a serum ALT flare exceeding fivefold the upper limit of the standard values after initiating TAF monotherapy.
Renal function and serum phosphorus levels in patients who switched from combination therapy with ETV plus TAF to TAF monotherapy
The median eGFR (mL/min/1.73 m2) at 0, 4, 12, 24, and 48 weeks was 66.3 (IQR 56.5–81.5), 72.2 (57.9–82.4), 68.4 (58.1–83.7), 66.2 (57.5–76.9), and 67.3 (54.1–79.0), respectively (Figure 2a). There was no significant change in eGFR after the initiation of TAF monotherapy. However, the changes in eGFR were −3.7 (−8.0 to −2.5) mL/min/1.73 m2 over the 48 weeks of TAF and ETV combination therapy, and by −2.1 (−3.8 to +3.1) mL/min/1.73 m2 over the 48 weeks of TAF monotherapy. Notably, the reduction in eGFR was significantly smaller during the TAF monotherapy period compared with the TAF and ETV combination therapy period (p = 0.0295, Wilcoxon signed-rank test; Figure 2b).
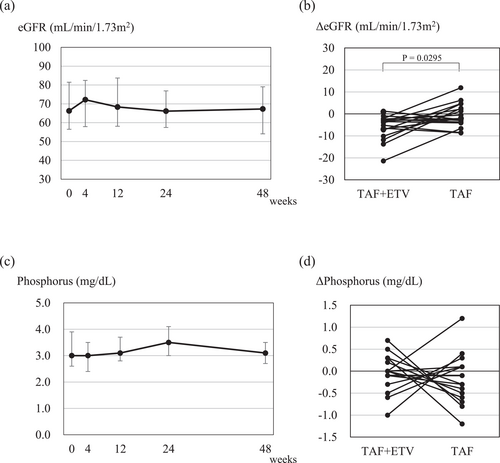
Renal function and serum phosphorus level in patients who switched from combination therapy with entecavir hydrate (ETV) plus tenofovir alafenamide fumarate (TAF) to TAF monotherapy. (a) Estimate glomerular filtration rate (eGFR) and (c) serum phosphorus levels at 4, 12, 24, and 48 weeks after switching to TAF monotherapy. The points and error bars represent the median values and quartiles (Q1 and Q3). (b) Changes in eGFR and (d) changes in serum phosphorus level during the 48-week TAF + ETV treatment period and 48-week TAF treatment period. The comparisons of changes in eGFR and phosphate levels were compared within the same patients.
Additionally, the median serum phosphorus level (mg/dL) at 0, 4, 12, 24, and 48 weeks was 3.0 (2.6–3.9), 3.0 (2.4–3.5), 3.1 (2.8–3.7), 3.5 (3.0–4.1), and 3.1 (2.7–3.5), respectively (Figure 2c). No significant changes were observed in the serum phosphorus level, and there was no significant difference in the changes between the TAF monotherapy period and the TAF and ETV combination therapy period (Figure 2d).
DISCUSSION
In the present study, we introduced an innovative method to identify drug-resistant mutations, encompassing minor clones, through direct and/or deep sequencing. This approach facilitates the analysis of HBV genome sequences even when serum HBV-DNA becomes undetectable during antiviral therapy with NAs. By utilizing this approach, the patients receiving combination therapy with ETV plus TAF for chronic HBV infection due to experiencing BTH or PR after initial LAM or ETV administration were able to safely transition to TAF monotherapy based on nucleotide sequences of HBV pregenome RNA in the sera. The monitoring serum HBV-DNA is available to confirm antiviral response and medication adherence during antiviral therapy. Cultured cells infected with HBV, as well as serum and tissue samples obtained through liver biopsy, show encapsulated RNA, which correspond to HBV pregenome RNA.11, 15 In the present study, however, not only encapsulated RNA, but also other forms of RNA, were extracted and subjected to the analysis. The achievement of HBV pregenome RNA amplification was associated with HBeAg, HBsAg, and HBcrAg levels. These observations supported that HBV pregenome RNA levels reflected the covalently closed circular DNA levels and HBV transcriptional activity under NAs administration. We examined in detail the drug-resistance profile of Pol/RT domain by direct sequencing and deep sequencing using HBV pregenome RNA from patients with persistent under detection sensitivity of serum HBV-DNA for a long period of time while receiving NAs. Furthermore, we reported for the first time the favorable antiviral response when switching to TAF monotherapy from the combination therapy of TAF and ETV in patients with chronic hepatitis B. Notably, we demonstrated that TAF monotherapy yields favorable viral response even in patients harboring drug-resistance mutations against LAM and/or ETV in the real-world cohort. This is because TFV shows no cross-resistance to LAM or ETV, demonstrating that TAF has high antiviral efficacy against HBV strains that are drug resistant to LAM and ETV in vivo.
The current Japan Society of Hepatology Guidelines for the Management of HBV Infection (4th edition) present the treatment strategy for good and poor responders to NA therapies in patients with chronic hepatitis B.16 In patients showing good response to ETV monotherapy, the guidelines recommend either continuing with ETV or considering a switch to TAF, taking into account the emergence of drug-resistant mutations. Furthermore, in patients showing poor response to LAM, ETV, TDF, or TAF monotherapy, the guidelines recommend changing the combination therapy with ETV and TAF. However, the guidelines lack a clear therapeutic strategy for patients currently undergoing the combination therapy of ETV and TAF. In the present study, we demonstrated that switching to TAF monotherapy from combination therapy with ETV and TAF is sufficient for maintain viral response in patients harboring drug-resistance mutations against LAM and/or ETV. Furthermore, five patients who did not show HBV-RNA amplification also dropped out of this prospective study, but TAF monotherapy was initiated based on the presumption of low transcriptional activity of the virus, and all of them are progressing well with good antiviral effects in TAF monotherapy. Although the sample size is limited, these findings imply that in clinical settings, patients who received ETV + TAF due to poor viral response against previous NAs might be transitioned to TAF monotherapy without necessitating HBV-RNA sequencing.
Recently, Park et al. reported that rtS106C, rtH126Y, rtD134E, and rtL269I show resistance to TFV. The IC50 against TFV of HBV strain harboring these quadruple mutations, as CYEI mutation, showed 15.3-fold higher than that of wild-type strain in vitro study.10 In contrast, Liu et al. reported that among 3886 patients with chronic hepatitis B enrolled in the clinical trial, two patients showed a CYEI mutation and one had a triple mutation of CYE, and these patients showed a good response to TDF or TAF monotherapy over 96 weeks.17 These observations suggested no significant clinical concerns about CYEI mutation. In the present study, none of the patients showed the quadruple CYEI mutation, and only one patient had the CEI mutation. Nevertheless, all patients with these mutations, including rtS106C, rtH126Y, rtD134E, and rtL269I, showed favorable response to TAF monotherapy.
It has been reported that combination therapies, such as LAM + ADV or ETV + ADV, have been effective in patients with poor response against ETV monotherapy.18, 19 Additionally, in a phase III trial of TDF in Japan, ETV + TDF combination therapy exerted a good response to patients who showed resistance to ETV monotherapy or ETV + ADV combination therapy. Furthermore, for patients with a resistance mutation against ETV, there was no significant difference in the antiviral response at the 48-week mark between the TDF monotherapy group and the TDF + ETV combination therapy group (71% vs. 73%).20 In the present study, among 18 patients showing poor response to initial therapy with LAM or ETV, three patients showed drug-resistance mutations to ETV and LAM, and four patients showed drug-resistance mutations to LAM. However, the antiviral efficacy remained favorable, even after switching to TAF monotherapy, in these patients. These observations provide scientific evidence for the effectiveness of TAF monotherapy against HBV strains harboring ETV resistance in real clinical practice.
Regarding drug-resistance mutations against TFV, a previous study reported that the coexistence of rtA194T with LAM-resistant mutations, such as rtL180M + rtM204V, in patients co-infected with HBV and HIV results in decreased susceptibility to TFV.8 In the present study, deep sequencing analysis detected coexistence of rtA194T with rtL180M, rtM204M, rtV173L, and rtT184A as 0.3% of minor clones in patient showing BTH during LAM administration. In this patient, TAF monotherapy showed a sustained antiviral response over a 48-week period. These observations revealed that rtA194T, as well as CYEI mutation, does not contribute to resistance to TAF.
In a long-term observation of 1298 patients who initiated TAF treatment enrolled in a phase III trial, no patient developed drug resistance mutation over 5 years, and the high resistance barrier of TAF was reported.21 In the present study, among 18 patients in whom HBV pregenome RNA was analyzed, a direct sequencing of the Pol/RT domain of HBV-DNA had been conducted for nine patents at a time when they showed a poor antiviral response to the initial therapy with LAM or ETV (a median of 8 years earlier), and nucleoside sequences of HBV-DNA at the time of viremia relapse and nucleotide sequences of HBV pregenome RNA during the combination therapy with ETV and TAF were found to be identical (data not shown). No amino acid substitutions, including drug resistance mutations, developed during the period of additional TAF administration after viremia relapse, confirming a high resistance barrier of TAF.
We previously conducted a prospective study on switching to TAF from ETV in patients who had achieved a good antiviral response.22, 23 That study showed a favorable antiviral response after switching to TAF. In that study, we conducted a questionnaire survey to assess medication adherence and satisfaction among patients who switched from ETV to TAF, because ETV must be taken on fasting stomach, whereas TAF can be taken regardless of meals. In the questionnaire survey, approximately 35% of patients mentioned that their dosing schedule had changed, approximately 30% reported a reduced frequency of forgetting to take their medication, and approximately 60% expressed satisfaction with the switching from ETV to TAF.22 Consequently, in our current study, the possibility of discontinuing ETV, which must be taken on an empty stomach, may have a positive impact on patient satisfaction with their medication.
In long-term administration of NAs, renal dysfunction often occurs due to mitochondrial impairment in the proximal renal tubules.24 In particular, the administration of ADV or TDF is associated with an increased risk of kidney damage, as evidenced by reduced eGFR and hypophosphatemia, necessitating close monitoring of renal function and serum phosphorus levels during prolonged use of these drugs.25 Conversely, there is minimal reporting of renal function impairment during long-term ETV administration. The advantage of ETV lies in its safety profile, especially in patients with pre-existing renal dysfunction. However, when administering ETV to patients with impaired kidney function, dose adjustment of ETV and continuous renal function monitoring are necessary. Although there is no report about renal toxicity directly attributed to ETV, it is possible that ETV could induce renal toxicity through various mechanisms, including renal tubular injury, apoptosis, and mitochondrial toxicity.25, 26 In the present study, the reduction in eGFR over 48 weeks after discontinuing ETV was significantly lower than that observed over the 48 weeks before discontinuation. These findings suggest that discontinuing ETV may contribute to the preservation of residual renal function. Furthermore, previous studies have reported an association between chronic HBV infection and chronic kidney disease.27-31 Specifically, it has been noted that glomerular diseases, such as membranous nephropathy and mesangiocapillary glomerulonephritis, could be underlying causes of renal dysfunction in patients with HBV infection.32, 33 Therefore, in the long-term management of chronic hepatitis B, it is imperative to closely monitor renal function. From this perspective, discontinuing ETV in patients on long-term administration with TAF and ETV may be beneficial for the patients. However, comparing changes in eGFR necessitates consideration of variables, such as age and alterations in muscle mass attributable to health conditions or sarcopenia. Therefore, further investigation is required to ascertain the renal benefits of transitioning from ETV + TAF to TAF monotherapy.
There were two limitations in the present study. The first limitation was the difficulty in proving the accuracy of the sequencing of HBV pregenome RNA. In the present study, HBV-DNA sequencing was not feasible, because all patients had HBV-DNA levels below the detection threshold. Consequently, we conducted only HBV-RNA sequencing. To mitigate these limitations, we confirmed that the sequences of HBV-DNA and HBV-RNA were consistent in nine patients at a time when they showed a poor antiviral response to the initial therapy with LAM or ETV. Additionally, in another cohort of patients with chronic hepatitis B before NAs treatment, we verified that the HBV-RNA sequences, when HBV-DNA became undetectable under NAs administration, aligned with the HBV-DNA sequences obtained before NA administration. These findings support the reliability of HBV-RNA sequencing in our study. The second limitation of this study was the small cohort size. Thus, it was not possible to establish a control group for comparing clinical outcomes, particularly antiviral effects, eGFR, and serum phosphate levels after ETV discontinuation. Given the limited number of patients, a comparison was conducted over a 48-week period before and after ETV discontinuation. In recent years, the occurrence of a drug-resistance mutation during NAs administration has become less frequent, making it difficult to conduct a study with a large number of patients within a single-center analysis. The findings of this study need validation in a multicenter larger cohort in the future.
In conclusion, it was possible to analyze drug resistance profiles in the Pol/RT domain of HBV by sequencing serum pregenome RNA in patients for whom serum HBV-DNA was undetectable during NAs administration. In patients developing drug-resistance mutations against LAM or ETV, the additional administration of TAF did not lead to the emergence of drug-resistance mutations against TAF. Furthermore, these patients showed a favorable antiviral effect, even when changing to TAF monotherapy. As a result, patients who are being treated with ETV plus TAF can safely change to TAF monotherapy.
CONFLICT OF INTEREST STATEMENT
Satoshi Mochida has received speaking fees or honoraria from AbbVie GK, Gilead Sciences Inc., Otsuka Pharmaceutical Co., Ltd., Toray Medical Co. Ltd., Eisai Co., Ltd. and ASKA Pharmaceutical Co., Ltd., has received research grants from EPS Corporation, Gilead Sciences Inc., MSD K.K. and intellim Corporation, and scholarship grants from AbbVie GK., EA Pharma Co. Ltd., Eisai Co., Ltd. and Sumitomo Pharma Co., Ltd. Nobuaki Nakayama is an Editorial Board member of Hepatology Research. The other authors declare no Conflict of Interests for this article
ETHICS STATEMENTS
Written informed consent was obtained from all patients. The study adhered to the ethical guidelines of the Declaration of Helsinki and was conducted with the approval of the Institutional Review Board of Saitama Medical University Hospital (2022-017). Additionally, the study was registered with the Japan Registry of Clinical Trials as jRCT1031220145.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.