BioTIME 2.0: Expanding and Improving a Database of Biodiversity Time Series
Correspondence:
BioTIME core Team ([email protected])
Funding: This work was supported by the H2020 European Research Council.
ABSTRACT
Motivation
Here, we make available a second version of the BioTIME database, which compiles records of abundance estimates for species in sample events of ecological assemblages through time. The updated version expands version 1.0 of the database by doubling the number of studies and includes substantial additional curation to the taxonomic accuracy of the records, as well as the metadata. Moreover, we now provide an R package (BioTIMEr) to facilitate use of the database.
Main Types of Variables Included
The database is composed of one main data table containing the abundance records and 11 metadata tables. The data are organised in a hierarchy of scales where 11,989,233 records are nested in 1,603,067 sample events, from 553,253 sampling locations, which are nested in 708 studies. A study is defined as a sampling methodology applied to an assemblage for a minimum of 2 years.
Spatial Location and Grain
Sampling locations in BioTIME are distributed across the planet, including marine, terrestrial and freshwater realms. Spatial grain size and extent vary across studies depending on sampling methodology. We recommend gridding of sampling locations into areas of consistent size.
Time Period and Grain
The earliest time series in BioTIME start in 1874, and the most recent records are from 2023. Temporal grain and duration vary across studies. We recommend doing sample-level rarefaction to ensure consistent sampling effort through time before calculating any diversity metric.
Major Taxa and Level of Measurement
The database includes any eukaryotic taxa, with a combined total of 56,400 taxa.
Software Format
csv and. SQL.
1 Background
The BioTIME database stores a curated collection of observations that can be used to estimate biodiversity metrics through time. Specifically, the database contains a collection of time series of observations of species abundances within biological assemblages that were sampled with consistent methods. With these data, it is possible to estimate temporal change in most metrics of taxonomic diversity (Magurran 2004), including, for example, species richness, evenness, and compositional change and population trends. We have assembled the database with the aim of facilitating synthesis studies and the re-use of these data by providing it in a standardised and curated format.
Since the publication of BioTIME version 1.0 (Dornelas et al. 2018), the database has been used for many different purposes. The first published analysis of the database revealed ubiquitous change in community composition, underpinned by roughly matched gains and losses of species through time (Dornelas et al. 2014). Other examples included the following: quantification of geographical variation in biodiversity change (Blowes et al. 2019; van Klink et al. 2020); estimation of the effects of temperature change (Antão et al. 2020), forest loss (Daskalova et al. 2020) and protected areas (Nowakowski et al. 2023) on biodiversity change; an estimation of the relationship between range shifts and population trends (Chaikin et al. 2024); and the quantification of change in organismal body size (Terry et al. 2021; Martins et al. 2023). Analysis of the BioTIME database also contributed one indicator to the first global assessment of biodiversity change produced by IPBES (2019).
In parallel with the proliferation of uses of BioTIME, the expansion and improvement of the database have continued. For BioTIME 2.0, additional dataset contributors were recruited, and updates were sourced for existing studies where data collection had continued. User feedback was also critical to flagging and resolving several inconsistencies not detected during the curation process of version 1.0. Moreover, metadata regarding methodology was updated and curation protocols were enhanced. In addition, the accuracy of taxonomic classification was checked and corrected where necessary. Finally, we developed a package in R (R Core Team 2023) to facilitate the usage of the database BioTIMEr (Sagouis 2024). We note that other databases have also been published with more focused criteria for inclusion (e.g., RivFishTIME focused on freshwater fish; Comte et al. 2020; InsectChange focused on insects; van Klink et al. 2021) or broader scopes (e.g., BioDeepTime which combines paleo and modern biodiversity time series; Smith et al. 2023). It is worth noting that there is only partial overlap between these databases and BioTIME because inclusion criteria differ across databases. For example, BioDeepTime includes only BioTIME time series longer than 10 years and combines these with multiple fossil databases. In addition, many studies in InsectChange did not meet BioTIME criteria for taxonomic resolution and/or lack of information on sampling methodology, which needed to be sourced independently. In summary, overlap among databases is nuanced, and care should be taken if combining BioTIME with other databases to avoid duplicate datasets.
Here, we release the updated version of BioTIME version 2.0. Given the twofold increase of studies in the database, the membership of the BioTIME consortium is also appropriately updated, as one of the goals of the database is to give credit to the data collectors.
2 Database Description
Similar to version 1.0, version 2.0 of the BioTIME database is a relational database composed of one main data table and 11 metadata tables. The data contained in the main table have a hierarchical structure (Figure 1): at which the finest scale is a record showing the observed abundance of a species; records are nested into sampling events, that is, a discrete moment in time and space when an assemblage is observed; a site is a location in space where one or more samples occur; multiple sampling events taken over time at the same site make up a time series; and time series are grouped into studies, which are defined by the sampling methodology, for example a specific type of transect with set length and width, or the trawl of a net of specified mesh size over a certain distance or length of time. Depending on the spatial study extent and the user definition of the grain size required for site, a study can have only one or multiple time series (see below in usage notes about the gridding process to define site).
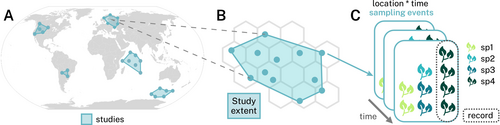
Metadata are stored in tables for: taxonomy (one table with taxonomy as provided and one table with standardised taxonomy), abundance type, biomass type, sample, study, methods, citation, contacts and curation. Only minor updates were done to the structure of the database relative to version 1.0, to accommodate additional taxonomic information (see below under Data curation and quality control and File S1 for a database schema which includes a description of the tables' fields).
3 Data Acquisition and Curation
New dataset acquisition for BioTIME 2.0 followed multiple approaches: active recruitment of data contributors in seminars, conferences and social media, searches for papers and within databases (e.g., OBIS [2024], GBIF [2024]), contributor volunteering, and through the collaboration networks of current data contributors. Once a candidate study was identified, it underwent checks against inclusion criteria and a curation process. For inclusion in BioTIME, studies must meet four criteria: (1) sampling methods are constant over time; (2) sampled for a minimum of two years, not necessarily consecutive; (3) samples take place at the assemblage scale rather than population; and (4) taxonomic resolution is mostly at the species level. We define a study as a single set of sampling or surveying methodology. If there are changes in methodology over time, candidate studies are split into multiple studies to reflect these changes, and split studies must independently fulfil BioTIME study criteria.
Once a candidate study was identified, available metadata and methodology information were used to build the metadata records (see protocol in File S2). Metadata records consist of information relating to temporal, spatial and taxonomic scope, habitat, methodology, protected area status, data originators and data sources. Where manipulation treatments were applied to some of the data, these were assessed as to whether the treatments were purely experimental manipulations (e.g., the artificial warming of a section), in which case only control samples were retained. If treatments were part of normal phenomena for the ecosystem (e.g., grazing), all samples were retained. Differences in ecological management practices were also recorded in the site metadata table to account for any differences in human activity/interactions.
Prior to inclusion in the database, data were standardised in our curation process. Quality control checks included checking for appropriate data types (e.g., numeric for abundance, string for species), realistic maximum and minimum values for fields, such as date and coordinates, removal of non-organismal records and correction of taxonomic misspellings, as per the taxonomic standardisation procedure described below. To store data in long format, records of null, blank or zeroes for abundances were removed; however, given the criterion that all species in the sample are recorded, absences can be interpreted as a species not being detected, and these can be reconstructed for each species in each time series.
Data standardisation also involved the construction of sampling event identifiers (‘SAMPLE_DESC’ in the raw data table). These are concatenated strings based on the provided study methods and data fields to accurately represent survey designs across space and time, such as sampling frequency and grouped observations (e.g., year_month_site_quadrat). The construction of these identifiers is reported in the metadata Sample table (‘SAMPLE_DESC_NAME’). The wide variety of sampling methods across the studies included in BioTIME is reflected in this field, with combinations of latitude, longitude, depth/elevation, date, transect, quadrat or trawl ID being common identifiers used. For some methods, for example, research cruise trawls, pitfall traps or camera traps, sampling was somewhat continuous. To represent the assemblage-level observations for these types of methods, samples were defined as constant time intervals (e.g., 1 week or 3 days depending on the nature of the data, but consistent within the time series). In the previous version of the database, we included a field to reflect whether observations took place in exactly the same location through time (e.g., in permanent plots), which has been deleted in this version of the database because of the difficulty in applying the concept consistently across taxa and methods (e.g., sessile vs. mobile taxa and destructive vs. observational data). Observation records are aggregated so that each sampling event contains only one abundance and/or biomass record per taxon, without any distinctions between life stage or sex, to ensure consistency across all datasets, and given that this was the resolution provided by the overwhelming majority of the studies. For studies added in BioTIME 2.0 where abundances and biomass are recorded at the individual level, records are not aggregated (i.e., abundance must be calculated by adding records of each species, and individual level sizes are kept within the database).
For version 2.0 of the database, records underwent a more rigorous standardisation of taxonomic classification. Specifically, all taxonomic records in the entire database were validated with either the taxize (Chamberlain and Szöcs 2013) or the worrms R packages (Chamberlain and Vanhoorne 2024). When using the taxize package, we used the classification() function and chose the Global Biodiversity Information Facility (GBIF) database as the first option to update the taxonomy, with the Integrated Taxonomic Information System (ITIS) as a second option should no matches be found. To ensure better representation of known marine species, we used the wm_records_names() function from the worrms package. We checked first for matches at the species level, then genus and, finally, family. If no valid names were found, we performed manual checks to the lowest resolution possible. Where species were identified as common names, we first ran them through the comm2Sci() function in taxize, before completing the checks as described above. BioTIME 2.0 contains two species tables: one which contains species as provided in the original data, and one with the standardised taxonomic classification, including species, genus, family, order, class, phylum and kingdom. Including the two tables ensures standardisation can be reproduced as taxonomy is updated. Nevertheless, it is worth noting that while lumping species that are synonymised is possible, splitting species beyond the data originally recorded is not.
BioTIME is designed to facilitate biodiversity analyses at the assemblage level, and hence any unidentified taxonomic records were kept to the lowest taxonomic resolution reported in the raw data. Records of unidentified taxa that were distinguished by the data collectors were kept separate (e.g., unknown beetle sp1, unknown beetle sp2) and are consistent within studies; therefore, these records can be used to estimate diversity metrics within the study, but cannot contribute to population assessments across studies (i.e., there is no way to determine whether populations of the same species appear in other studies). The standardised version of the database has 97% of the taxa identified to at least family and 74% to species level.
For spatial information, latitudes and longitudes of each study were mapped to check they matched location descriptions. Spatial extent was estimated as the area of the convex hull encompassing all the spatial coordinates (Figure 1) and grain size from the reported methods for each study. Changes made during the curation process were recorded in the curation table and confirmed with the data providers. For all studies revised or added to this database version, code used in data curation is available from the BioTIME Github repository (https://github.com/bioTIMEHub/BioTIME). The curated version of the data was shared with the data providers who agreed to the changes made.
In this version, 16 studies previously included in BioTIME v1.0 were removed, as additional information revealed they did not meet some of the criteria for inclusion in the database (File S4). Additionally, 49 studies included in BioTIME v1.0 were recurated as more metadata or new data became available—all these changes are reported in File S4.
The contact table includes publicly available contact information for data contributors (name and/or email) to allow users to reach out to the original contributors with any queries regarding data usage. These data were processed in compliance with both UK and EU General Data Protection Regulations (GDPR). A data protection statement explaining the lawful basis for the use and processing of these data is now available on the database website: https://biotime.st-andrews.ac.uk/usageGuidelines.php.
4 Description of Data
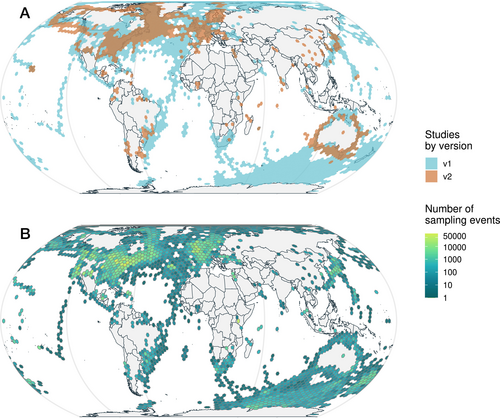
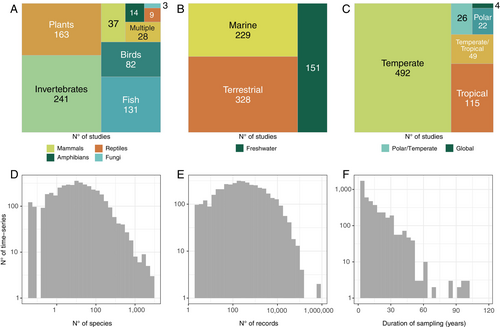
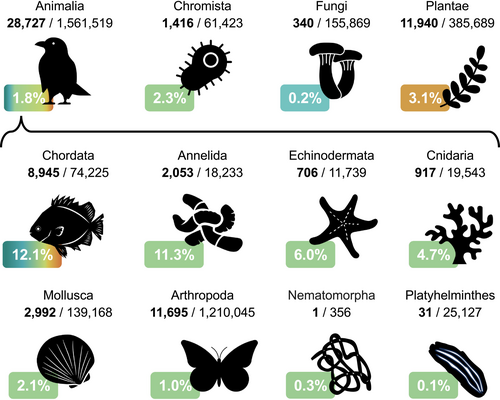
BioTIME 2.0 includes 708 studies distributed across 553,253 locations, with almost twice as many studies and 11.3% more locations relative to the previous version (Figure 2). The database now includes 11,989,233 records from 56,400 taxa (36.7% and 26.7% increase from version 1.0, respectively) from across the tree of life, collected over 1,603,067 sampling events across the marine, freshwater and terrestrial realms (Figure 3). Temporally, the database spans 1874 to 2023, with median time series length being 7 years. With a grid resolution of 75,000 km2, the database currently includes 4,301 time series in total, of which, 2390 have durations longer than 5 years, 1,745 longer than 10, 893 longer than 20, and 37 longer than 50 years (Figure 3). Despite efforts to improve representation, both spatial and taxonomic biases persist (Figures 2 and 4, File S5). Spatial biases persist in the database and are especially evident in the terrestrial realm, despite targeted searches having improved spatial representation. The marine realm has better representation, both spatial (in terms of latitudes and longitudes) and regarding global change space (Daskalova et al. 2020). However, as inherently more three-dimensional and given the features of sampling in marine habitats, it is likely that a smaller proportion of the marine realm is represented in our database compared with the terrestrial realm.
5 Usage Notes
This version of the database is made publicly available in a SQL version and as a .csv query through Zenodo (10.5281/zenodo.10932823) and BioTIME's website (https://biotime.st-andrews.ac.uk) under a CC-BY licence (https://creativecommons.org/). The data are, hence, free to use with attribution via citation of this paper. In addition, each study has a licence associated with a spectrum of governmental, Creative Commons and Data Commons licences. The database is also GDPR compliant. Citations for data sources of individual studies are provided in the metadata table citation and are also listed in File S3.
To facilitate comparisons across studies, we recommend standardising the spatial extent of the time series by gridding the data into constant area polygons prior to analysis. In addition, as the number of samples may change through time, we recommend sampling effort standardisation. To facilitate the use of the database, the release of BioTIME 2.0 is accompanied by an R package, BioTIMEr (Sagouis 2024). The package provides functions to deal with these spatial and temporal issues—namely to spatially grid the studies into constant extent cells and subsample time-series so that sampling effort (specifically number of samples) is constant through time. In addition, the package includes functions to calculate several metrics of alpha diversity and compositional change over time. A vignette is supplied to illustrate the use of each function.
The extended efforts in data standardisation aimed to facilitate integration with other databases. For example, the taxonomic standardisation should streamline integration with trait or phylogenetic data, and for this purpose, the standardised species name is preferable. In contrast, to reflect the species names as recognised by the observers at the time of observation, or to update as taxonomy changes, the original species names are preferable.
Acknowledgements
The BioTIME project was funded since conception by the European Research Council grant AdG BioTIME 250189 awarded to A.E.M., European Research Council grant PoC BioCHANGE 727440 awarded to A.E.M. and M.D., European Research Council grant AdG MetaCHANGE 101098020 awarded to J.C. and the Leverhulme Centre for Anthropocene Biodiversity grant RC-2018-021 awarded to M.D. These funds partially supported V.B., C.C., A.F.E. and F.M. A.F.E. was supported by the Fisheries Society of the British Isles Studentship. V.B. and M.D. were partially supported by European Research Council grant CoralINT 101044975. Views and opinions expressed are, however, those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council. Neither the European Union nor the granting authority can be held responsible for them. We would like to thank Ramon Fallon and Swithun Crowe for their assistance in web maintenance. The BioTIME Core Team would like to thank Amelia Penny, Anokhi Saha and Rowan Stanford for their help in data curation. We would also like to thank the following individuals for contributing and collecting data: Alejandro Pérez Matus, Amir Hosain Chowdhury, Anders Enemar, Andrew Rassweiler, Andy Kirk, Anna Maria Fosaa, Anthony Joern, Anu Valtonen, Anywa Waite, A.P. Savassi-Coutinho, Arik Diamant, Asem A. Akhmetzhanova, Atsuko Fukamachi, B. R. Ramesh, Beatriz Eugenia Salgado Negret, Bella Galil, Bill McLarney, Borgþór Magnússon, Brian J. Bett, Bruno Ferreto Fiorillo, Cameron Eckert, Carol Mathias, Christopher C. Koenig, Conor Waldock, Corinna Gries, Dan Bardsley, Dan Childers, Dan Metcalfe, Dave Watts, David Janik, David Lightfoot, David Rissik, Diego Montalti, Don Henshaw, Douglas Kelt, Dustin Wilgers, Elizabeth Gorgone, Ellen Marshall, Erica M. Sampaio, Esther Levesque, Even Moreland, Fábio Lang da Silveira, Fakhrizal Setiawan, Fernando Carvalho, Fernando Valladares, Fernando Vaz-de-Mello, Francisco Sánchez-Piñero, G. A. D. C. Franco, Gaius Shaver, Gedas Vaitkus, Grace Frank, Grace Murphy, Haley Arnold, Hannah Ruhl, Hanneline Smit-Robinson, Haydn Thomas, Heather L Bateman, Hernando García, Ian Douglas-Hamilton, J. Y. Tamashiro, James H. Brown, Jan Clavel, Javier Bustamante, Jennifer Wallace, Jenny Owen, Jill Johnstone, Jo Surgey, John C. Priscu, John F. Chamblee, John R. Clark, Jose Manuel Arcos, Josh Silberg, Julio Gutiérrez, Kamil Kral, Kathy Slater, Keith Stedman, Kwang-Tsao Shao, Laura Siegwart Collier, Lenka Kočíková, Lesley Clementson, Linda A. Kuhnz, Loureiro Fernandes, Malva Medina, Marc Estiarte, Marco Moretti, Marion Valeix, Mark Harmon, Mark Ritchie, Mark Watkins, Martin Schuetz, Masato Yoshikawa, Matt Bradford, Michelle Devlin, Mike Brook, Mike Brubaker, Miles Furnas, Nadia Soudzilovskaia, Natalia Norden, Ngoni Chiweshe, Nir Stern, Oliver Beacock, Olivia Mendivil Ramos Or Givan, Orjan Totland, Paul Richardson, Pedro Damião dos Santos Rebelo, Pedro José Sandoval Cortés, Peter Adler, Peter Kirmond, Phaedra Budy, René E. Maragliano, Renee Patten, Res Altwegg, Richard Dufeu, Rita Adrian, Rob Pabst, Robert Fitt, Robin Elahi, Roger Hnatiuk, Sally Sherman, Sandra Angers-Blondin, Shahar Malamud, Sharon Collinge, Shaun Wilson, Sophie Leterme, Stefan Williams, Stephen Hale, Stephen Newton, Steve Hubbell, Steve Wain, Sukmaraharja Aulia Rachman Tarigan, Sultan Areshi, Susan Boyd, Susannah B. Lerman, Susi Pulliainen, Tae-Sung Kwon, Tasrif Kartawijaya, Thomas Wohlgemuth, Tiago Egydio Barreto, Tim S. Doherty, Tomáš Vrška, Tomasz Weslowski, Tomohiro Yoshida, Tore Johannessen, Tracey Smart, Trevor Willis, Tung-Yung Fan, Vinícius Castro Souza, Vu Quang Con, Ward Appeltans, Ya'arit Levitt Barmats, Yoshinobu Hoshino and Yoshiyuki Umatani. The dataset by A. Barash was collected as part of the research activity of Sharks in Israel NGO. AT was supported by JSPS KAKENHI (grant no.: 20H03010). A.F. would like to thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 312689/2021-7) for a productivity grant and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2015/06743-0) for a grant. A.K. was supported by EXPRO (grant no.: 19-28807X) (Czech Science Foundation) and a long-term research development project RVO 67985939 (Czech Academy of Sciences). Funding for A.V. Christianini was provided by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP proc. no. 02/12895-8); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)—Finance Code 001; Neotropical Grassland Conservancy; and Rufford Small Grants. The Zimbabwe Parks and Wildlife Management Authority and the Zimbabwean Veterinary Services kindly granted A. Caron permission to work in areas under their jurisdiction. This work was conducted within the framework of the ‘Mesures d'Urgence’ and GRIPAVI projects, and the Research Platform ‘Production and Conservation in Partnership’ (RP–PCP). A.H.H. was funded by the National Science Foundation (grant no.: DEB-1354563). Thanks to Devan Inderlall, Khadija Huggins and Sarah Shageer for assistance processing macroinvertebrate samples, and to Avinash Deonarinesingh for fieldwork assistance. Thanks to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for granting a research productivity scholarship to Ana Carolina da Silva. The study by A. Michelsen was supported by Independent Research Fund Denmark, D.F.F. Nature and Universe. This manuscript uses data collected by the US National Science Foundation's (NSF) Moorea Coral Reef Long-Term Ecological research (MCR LTER) site (grant no. OCE 2224354) (and earlier awards). Research was completed under permits issued by the French Polynesian Government (Délégation à la Recherche) and the Haut-commissariat de la République en Polynésie Francaise (DTRT) (Protocole d'Accueil 2005-2023). A.L.R. was supported by the California Agricultural Experimental Station of the University of California Davis (grant no.: CA-D-WFB-2467-H). This work was supported by the UK Natural Environment Research Council (NERC), through the Climate Linked Atlantic Sector Science project (NE/R015953/1). M.B., A.M.J., K.P. and J.S. received financial support from internal funds of University of Lodz. Trinidad data from A.E. Magurran and A.E. Deacon were supported by the European Research Council (ERC AdG BioTIME 250189) and by the Leverhulme Trust (RPG-2019-402). A. Thimonier would like to thank these botanists for their assistance in the Swiss LWF data set: Peter Kull, Walter Keller, Thomas Wohlgemuth, Barbara Moser, Martin Schütz. A. Tolvanen would like to thank Natural Resources Institute Finland. Data on plankton abundance in Australia were sourced from Australia's Integrated Marine Observing System (IMOS)—IMOS is enabled by the National Collaborative Research Infrastructure Strategy (NCRIS). It is operated by a consortium of institutions as an unincorporated joint venture, with the University of Tasmania as Lead Agent. Funding to A. Bode was provided by projects RADIALES (IEO-CSIC) and QLOCKS (PID2020-115620RB-100 MCIN/AEI/10.13039/501100011033, Spain). The study by A. Verstraeten was funded partially by the UNECE and partly by the Flemish government. A.V. is also grateful to Luc De Geest for assisting with the data collection in the field. The surveys by A. Zvuloni were made possible thanks to funding from the Israel Nature and Park Authority. We wish to thank all those who helped with fieldwork and the Interuniversity Institute for Marine Sciences in Eilat (IUI) for its support. B.P. Nikolov: Public data provided by the Executive Environment Agency (ExEA), of the Ministry of Environment and Water, Bulgaria. Original data have been provided/collected by Wetlands International, as well as by specialists and experts from ExEA, RIEW, some research organisations and NGOs within the monitoring of wintering waterbirds as a part of the Bulgarian National Monitoring System for the condition of biodiversity. Data collected at the Jornada Experimental Range were supported by the USDA Long-Term Agroecosystem Research network and Jornada Basin Long-Term Ecological Research Program. B.F.-Z. was supported by an Abroad Doctoral Scholarship ANID (National Research and Development Agency), Chile. B.M.S. was supported by the Natural Science and Engineering Research Council of Canada, Hakai Institute, BC Parks. C.Z.T was supported by the Brazilian Federal Agency for Support and Evaluation of Graduate Education (CAPES #001). C. Gjerdrum would like to acknowledge the financial support (2006-2009) of the Environmental Studies Research Funds (ESRF, http://www.esrfunds.org), and grants US NSF 1022472 and 1902595 for ship time support in Davis Strait/Baffin Bay in 2015. Thanks to all the observers who participated in collecting the data, as well as science staff, ships' officers and personnel for support at sea, especially the Department of Fisheries and Oceans in eastern Canada. We thank Jan Wittoeck and other colleagues who assisted in the sampling and compilation of the macrobenthic data and the Belgian Federal Science Policy Office who funded MACROBEL through the programme ‘Sustainable management of the North Sea’ (SPSD I MN/02/96). C.A.H.F. would like to thank FAPESP for its financial support of part of the data gathering and analysis for a number of years. Funding to C.E.B. was provided by the Bertarelli Foundation as part of the Bertarelli Programme in Marine Science. Funding for C. Rodrigues was provided by Conselho Nacional de Desenvolvimento Científico e Tecnológico (grant/award no.: 312689/2021-7); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (grant/award no.: 001); Fundação de Amparo à Pesquisa do Estado de São Paulo (grant/award nos.: 2015/06743-0 and 2020/10333-0); Fundação Grupo o Boticário de Proteção à Natureza (grant/award no.: 0153_2011_PR). Data from microzooplankton from the Bahía Blanca Estuary have been gathered and processed jointly by Dr. Sonia Barría de Cao, Dr. Rosa Pettigrosso and Celeste López-Abbate. C.A.C. would like to thank Academia Sinica, National Science and Technology Council, and Kenting National Park for funding. CHH would like to thank the National Science and Technology Council, Taiwan. Funding to C.L.H. was provided by David and Lucile Packard Foundation. This work was made possible by the RV Western Flyer crew, and pilots of ROVs Tiburon and Doc Ricketts. Funding to C.F.J.M. was provided by a Fundação para a Ciência e a Tecnologia grant (PTDC/BIABIC/111184/2009). Funding to C.D.S. was provided by a grant from the US Fish and Wildlife Service/State Wildlife federal grant number: T-15, Florida Fish and Wildlife Conservation Commission agreement number: 08007. We thank A. Mickle and B. Hall-Scharf for assistance in the field and the laboratory, and M. McManus for advice on study design. Claire Widdicombe and the L4 phytoplankton time-series are funded by the UK's Natural Environment Research Council's National Capability Long-term Single Centre Science Programme, Climate Linked Atlantic Sector Science (grant no.: NE/R015953/1), and is a contribution to Theme 1.3—Biological Dynamics. Data from C.H.D. were sourced from Australia's Integrated Marine Observing System (IMOS)—IMOS is enabled by the National Collaborative Research Infrastructure Strategy (NCRIS). It is operated by a consortium of institutions as an unincorporated joint venture, with the University of Tasmania as Lead Agent. Data from D.A.E. were collected thanks to logistical support and funding from Operation Wallacea. D.C.R. and R.J.M. thank the US National Science Foundation for their ongoing support in funding the Santa Barbara Coastal Long-Term Ecological Research (SBC-LTER) Program. D.K.S. thanks the dozens of researchers who sampled crayfish, macrophytes and snails in northern Wisconsin, USA, throughout the duration of the study, including Timothy A. Kreps, T. Mark Olsen and Sadie K. Rosenthal. D. Oro thanks the Spanish Ministry of Science and Generalitat de Catalunya for funding and support. D.J.K. and colleagues acknowledge Channel Islands National Park, the many park and volunteer divers, as well as the National Park Service Inventory and Monitoring Program for continued support of the long-term Kelp Forest Monitoring Program. D.M.L. thanks the US NSF, Wisconsin DNR, Cornell Atkinson Center for Sustainability. D. Maphisa: The first author was supported in the position of BirdLife South Africa Ingula Project Manager with funding by Eskom through The Ingula Partnership and by The Mazda Wildlife Fund with a vehicle for the duration of the project while employed by BirdLife South Africa. D.C.R.-F. is funded by CNPq #304760/2021-8. Funding for DFF's fieldwork was provided by the Madeira 420 European Regional Development Fund Programme management authority under the Regional Government of Madeira. D.F.F was supported by Scientific Initiation Studentship (RH015). Funding to D.L.F. provided by US NSF, NOAA and NASA. D. Edelist acknowledges the Department of Fisheries, Israel Ministry of Agriculture and Rural Development. E.C.-M., V.H.R.-M. and R.R.T. thank NSF-Florida Coastal Everglades Long-Term Ecological Research (FCE-LTER) Program (grant nos.: DBI-0620409, DEB-9910514 and DEB-1237517). Funding for E.J.C. came from the Norwegian Research Council (‘SnoEco’ project, number: 230970), European Commission ‘FRAGILE’ project (no.: EVK2-2001-00235), UiT—The Arctic University of Norway and The University Centre in Svalbard (UNIS). The SOTS observatory is supported by Australia's Integrated Marine Observing System (IMOS). IMOS is operated by a consortium of institutions as an unincorporated joint venture, with the University of Tasmania as Lead Agent. Konza Prairie LTER: NSF DEB #2025849. E.G.B. mostly received financial support from Fundació Rivus, Observatori de la Tordera and the Spanish Ministry of Science (MCIN/AEI/ 10.13039/501100011033) and the European Union (NextGenerationEU/PRTR) (project TED 2021-129889 B–I00). North Temperate Lakes LTER datasets were funded by the National Science Foundation under Cooperative Agreement #DEB-2025982. E.P. acknowledges support from the US National Science Foundation under awards 0217259, 0713994, 0902125, 1107381 and 1525636. This research was supported by appropriated funds to the USDA-ARS Jornada Experimental Range project 3050-11210-009-00D, the Jornada Basin Long-Term Ecological Research Program DEB-1832194, and is a contribution from the Long-Term Agroecosystem Research (LTAR) network. LTAR is supported by the United States Department of Agriculture. Peter Adler was supported by NSF DEB-0614068 and DEB-1054040 and the Utah Agricultural Experiment Station. E.M.C. also wants to thank the many personnel who contributed to data collection over the Jornada quadrat programme's 100-year history, with special thanks to Kris Havstad for reviving the project in 1995. The data obtained at Sapucuá lake are a secondary product of the project Ecological studies in the low-order streams in the National Forest Saraca-Taquera (PA), headed by Francisco de Assis Esteves and Reinaldo Luiz Bozelli, supported by Mineração Rio do Norte. We are grateful to all participants of the monitoring by help with fieldwork and curation of biological samples. E. Bonsdorff thanks The Åbo Akademi University Foundation. The work by E.C.K. was supported by the National Science Foundation's McMurdo LTER project. Support was also supplied by INSTAAR (Institute of Arctic and Alpine Research of the University of Colorado, Boulder) and was directed by Dr. Diane McKnight. The work by E.P.W. was supported by the USDA National Institute of Food and Agriculture, Hatch project FLA-WEC-005944. F.V. would like to thank Fazenda Vagafogo. FRdS thanks the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP #2013/50714-0 and #2022/04012-2). F.C.S. was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo—FAPESP (Filipe C. Serrano, grant no.: 2023/06999-1). F.A.M.S. was supported by CNPq 310168/2018-0; FAEPEX 2012/23. The research of F.P.D. was funded by NSF grants DEB-1237733 and NSF 0080381 awarded to the University of Virginia in support of the Virginia Coast Reserve LTER site. F. West would like to acknowledge Jonathan West, late principal of the Whitelands Project, as he was instrumental in gathering this dataset. G.B.G.S. and M.V. thank the staff who assisted in the field and laboratory research from the Laboratory of Fishery Biology and Technology. This study was part of the programme ‘Environmental Assessment of Guanabara Bay’ coordinated and funded by CENPES—PETROBRAS, which has given permission for the publication of the results. This study was also supported by the Long-Term Ecological Programme (PELD programme—CNPq 403809/2012-6) and by FAPERJ (Thematic Programme, process E-26/110.114/2013). G.B.G.S. was funded by CAPES/Brazil. G. Damasceno was supported by FAPESP (São Paulo Research Foundation) (grant/award nos.: 2015/10714-6, 2015/06743-0). The ILVO data collection was funded through the Bagger project (Biological and chemical ecosystem monitoring at the dredge disposal sites financed by Flemish Authorities, Flanders Maritime Access Division); Zand (Financial support was provided by the continuous monitoring programme ZAGRI, paid from the revenues of marine aggregate extraction in Belgian waters (RD 9/04/2014, Numac: 2014011344) and sampling was possible through the RV Belgica (BELSPO funding). G. Durigan thanks the Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq (grant no.: 309709/2020-2). The research by G.S.C. was partially supported by The Wildlife Conservation Society through funding provided by US-AID for the Global Avian Influenza Network for Surveillance. Long-term ecosystem research in the Scheldt Estuary (Belgium) by MONEOS programme is supported by ‘De Vlaamse Waterweg nv’ and ‘Maritieme toegang’. Funding to H. Knutsen was provided by the Ministry of Trade, Industry and Fisheries. H.M.V. thanks the staff of various South African national government departments historically and currently involved in the collection and analysis of zooplankton samples during routine fisheries and environmental monitoring surveys in the South-East Atlantic along the west coast of South Africa. H. Bruelheide appreciates the support for the strategic project sMon by the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, funded by the German Research Foundation (DFG-FZT 118, 202548816). H.J.W.V. extends thanks to the Dutch nature conservation organisation Natuurmonumenten for contributing to the costs and Saskia Glas for the support. LTER ICP Solling Germany Beech Forest Ground Vegetation, LTER ICP Solling Germany Spruce Forest Vegetation: The execution of the relevés by Bernadett Lambertz is gratefully acknowledged. Funding was provided through the Permanent Soil Monitoring programme of Lower Saxony. The study by H. Taki was partly funded by the Joint Research Program of the Institute of Low Temperature Science, Hokkaido University, the Global Environment Research Fund of the Ministry of Environment of Japan, and the Ministry of Agriculture, Forestry and Fisheries of Japan. H.T.P. thanks Fundação de Amparo à Pesquisa do Estado de São Paulo (grant nos.: 2019/24215-2 and 2021/07039-6). The Finnish Moth Monitoring scheme (Nocturna) has been supported financially by the Ministry of the Environment (Finland). I. Torre and colleagues are indebted to Diputació de Barcelona and the Collserola Natural Park for providing financial and logistic support throughout the years. The authors thank Antoni Arrizabalaga (laboratory's head) for providing bureaucratic support and guarantee to the SEMICE programme throughout the years. We also acknowledge the volunteers and professionals in charge of the SEMICE stations, who kindly recorded data in the study area: Lídia Freixas, Tomàs Pulido, Dolors Escruela, Òscar Martínez, James Manresa, Cristina Terraza, Joan Manuel Riera, Marçal Pou, Tabea Sunnemann and Alfons Raspall. IdCA and colleagues thank Prof. Guy Chavanon for obtaining the sampling permits, and the Moroccan Direction de la Lutte Contre la Désertification et de la Protection de la Nature for permission to conduct this research (Refs. 01/2013 HCEFLCD/DLCDPN/DPRN/CFF and 01/2014 HCEFLCD/DLCDPN/DPRN/CFF). Several researchers, graduate and postgraduate students helped during field campaigns. I. deCastro-Arrazola was funded by a FPI grant from the Spanish Ministry of Science and Innovation (BES-2012-054353). Surveys and data processing were supported by the Spanish Agency of Innovation (AEI) project SCARPO (grant no.: CGL2011-29317). I.S.M. was funded by Marie Sklodowska-Curie Actions Individual Fellowship (MSCA-IF), European Union's Horizon 2020 (grant agreement no.: 894644); Leverhulme Trust through the Leverhulme Centre for Anthropocene Biodiversity (RC-2018-021). The work by I.K.S. was supported by eLTER-PLUS and LTER-DK projects. I.H.M.-S. was supported by the Natural Environment Research Council of the UK (NERC), European Research Council (ERC) and Canada Excellence Research Chairs Program funded by the Natural Sciences and Engineering Research Council of Canada (NSERC). For the ITEX data collected on Herschel Island-Qikiqtaruk, we thank the Herschel Island-Qikiqtaruk Territorial Park management, Catherine Kennedy, Dorothy Cooley and Jill F. Johnstone for establishing and maintaining the plant composition data from Qikiqtaruk. We thank the Inuvialuit People for the opportunity to conduct research on their land. Data and facilities to I. Arismendi and D. Bell were provided by the H.J. Andrews Experimental Forest and Long-Term Ecological Research (LTER) Program, administered cooperatively by the USDA Forest Service Pacific Northwest Research Station, Oregon State University and the Willamette National Forest. This research is based upon work supported by the National Science Foundation under the LTER Grants: LTER7 DEB-1440409 (2012-2020) and LTER8 DEB-2025755 (2020-2026). Data from J.E.D. are Contribution 141 from the Smithsonian's MarineGEO and Tennenbaum Marine Observatories Network. Field support to J.W.F.C. came from Ocean Networks Canada, captains and crew of the CCGS JP Tully, CCGS Vector, RV Thompson, the RV Falkor, and staff of the Canadian Scientific Submersible Facility and operators of the Oceanic Explorer. Funding came from NSERC and the Canadian Healthy Oceans Network (CHONe). J.A.S. acknowledges funding support from the National Research Foundation of South Africa (grant nos.: 150296, 118593 and 142438). J. R. S. Vitule is especially thankful to the Brazilian Council of Research (CNPq) for continuous funding through Research Productivity Grants. J.P.S.V.A. was supported by FAPESP Processo 2020/12658-4 AND FAPESP Processo 2023/01470-9. R.K., D.P. and J.M. acknowledge and thank SOTEAG (Shetland Oil Terminal Environmental Advisory Group) for providing data from the long-term rocky shore monitoring programme dataset. J.M., D.P. and R.K. would like to acknowledge Christine M. Howson for her considerable contribution to the SOTEAG rocky shore programme over many years. J.J.L. acknowledges funding from the Research Foundation Flanders (FWO, project nos.: G018919N, 12P1819N, W001919N and 1512720 N). J.J.L. also acknowledges BiodivERsA project ASICS (G0H6720N) and funding from INTERACT travel grants. J.M.C. acknowledges the support of iDiv funded by the German Research Foundation (grant nos.: DFG-FZT 118, 202548816). Ainsdale Dunes Slacks LTE is hosted by Natural England and received funding from the Ecological Continuity Trust (ECT), the Botanical Society of the Britain and Ireland (BSBI), Loughborugh University and the Leverhulme Trust as part of the award of a Royal Society Leverhulme Trust Senior Research Fellowship to Millett. J. Barlow thanks the UK Government Darwin Initiative. J. Penuelas was supported by Spanish government grant PID2022-140808NB-I00. J.C.Q. thanks the German Research Foundation (DFG) granted to the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig (DFG FZT-118, 20254881). Data from J.P.Q. are a contribution of the Research Center for Marine Biodiversity of the University of São Paulo (NPBiomar). The Finnish Moth Monitoring scheme (Nocturna) has been supported financially by the Ministry of the Environment (Finland). Funding for J.A.J. was provided by National Science Foundation grants to the Andrews Forest Long-Term Ecological Research Program. K.E.E. and colleagues acknowledge Offshore Norge for use of data from the MOD-database. K. Klanderud thanks the Norwegian Research Council. Dr. Fiona Scott is acknowledged for her excellent work with the SO-CPR programme. Data collection for K.D.W. assisted by > 30 Bennington College undergraduates; researchers involved in pre-1989 censuses are cited with datasets archived at Environmental Data Initiative. K. Meier was supported by workgroups Research Data Management and Data Infrastructures at ZALF. L.H.A. acknowledges funding from the Academy of Finland (grant no.: 340280). Living data internship was provided by Laura Super, PhD Candidate, University of British Columbia, with the assistance of the team wrote up the first draft of this Readme file and did data file curation—including making data summary files and completing data cleaning—as part of a Living Data Project (LDP), Canadian Institute of Ecology and Evolution, data rescue paid internship supported by NSERC CREATE (https://www.ciee-icee.ca/ldp.html). In addition to the team, there was cohort support from LDP graduate students and postdocs. See more here about the principles and rationale behind data rescue: Bledsoe, E. K. et al. (2022). Data rescue: saving environmental data from extinction. Proceedings of the Royal Society B, 289(1979), 20220938. https://doi.org/10.1098/rspb.2022.0938. Funding to LTT was provided by two UK Natural Environment Research Council PhD studentships (including through the Quadrat Doctoral Training Program) and a PhD studentship funded by the Saudi Royal Embassy. L.V.V. was supported by the Vietnam Ministry of Science and Technology (ĐTĐL.CN-113/21) and Earthwatch Institute. The dataset by L. Armbrecht originated from PhD research, which was supervised by Leanne Armand, Moninya Roughan and David Raftos. We thank the NMSC for providing RV Circe and the OEH for enabling the use of RV Bombora. We thank P. Ajani and T. Ingleton for support with phytoplankton identification. Throughout the many field campaigns, we thank T. Ingleton, P. Davies, R. Gardiner, A. Cox, B. Morris, D. Kricke, A. Waite, T. Sullivan, V. Rossi, J. Wood, T. Austin, M. Doblin, C. Robinson, A. Mantovanelli, A. Schaeffer, A. Grobler, W. Godinho, A. Wilkins and S. Milburn for assistance. The field work was partially funded by a grant from the Australian Research Council, DP 1093510 (to MR). L.A. was funded by Macquarie University, the Australian Biological Resources Study, the Australian Marine Sciences Association and the Linnean Society of NSW. IMOS is supported by the Australian Government through the National Collaborative Research Infrastructure Strategy and the Super Science Initiative. L.B.C. acknowledges the contribution by the European Union's Horizon 2023 Research and Innovation Program under grant agreement no.: 101060072 ACTNOW and grant agreement no.: 101058956 Marine SABRES. RivFishTIME has been developed as part of the international working group sYNGEO—The geography of synchrony in dendritic networks kindly supported by sDiv, the Synthesis Centre of the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, funded by the German Research Foundation (FZT 118). L. Rosselli and colleagues thank the Asociación Bogotana de Ornitología and all observers that participated in the Sabana de Bogotá Christmas Bird Counts. We also thank landowners and institutions that permitted access to the areas during the decades of CBC. Juanita Niño, Julián Pinzón and Stefany Velásquez-Licona of the UDCA helped with organising the data. For the Chingaza study, we are grateful to the Sistema de Parques Nacionales Naturales de Colombia for help with logistics and fieldwork and for granting permits. The Empresa de Acueducto de Bogota helped with logistics in 1991–1992. For companionship and valuable help in the field, we thank P. Camargo, F. Avellaneda, J. Candil, L. G. Linares, N. Moreno, R. Pulido, L. Tellez and the students of the UDCA. C. Garzon-Sanabria helped with data organisation. We thank G. Castañeda for his enthusiastic support in transportation in the field. The Chingaza project was funded by Universidad de Ciencias Aplicadas y Ambientales UDCA, Universidad Nacional de Colombia and resources from Patrimonio Autonomo Fondo Nacional de Financiamiento para la Ciencia, la Tecnologıa y la Innovacion, Francisco Jose de Caldas. L. Crevecoeur thanks Marc Janssen. Research Productivity Grant (CNPq) (process #302881/2022-0). Grants and fellowships were provided to L.F.T. by São Paulo Research Foundation (FAPESP #2022/11096-8) and the National Council for Scientific and Technological Development (CNPq #302834/2020-6). We thank ArcticNet, Weston Northern Grants. M. Lazarus and colleagues would like to thank the Israeli Nature and Park Authority and all present and past Belmaker laboratory members who participated in data collection. M. Kagami was funded by JSPS_KAKEN(16H02943). M. Vianna acknowledges FAPERJ (E26/200.934/2022) and CNPq (302398/2022–8) for financial support. Marcio Martins thanks the Fundação de Amparo à Pesquisa do Estado de São Paulo (Fapesp) for research grants (#2015/21259-8, #2018/14091-1 and #2020/12658-4) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for research fellowships (#306961/2015-6 and #309772/2021-4). M. Schaub: Evaluations were based on data from the Swiss Long-term Forest Ecosystem Research programme LWF (www.lwf.ch), which is part of the UNECE Cooperative Programme on Assessment and Monitoring of Air Pollution Effects on Forests ICP Forests (www.icp-forests.net). We are particularly grateful to Tom Wohlgemuth who helped assess the Swiss data. M.G.C. was funded by the EU Horizon 2020 Research and Innovations Programme through the CHARTER project (grant no.: 869471) and by the NERC Tundra Time project (NE/W006448/1). M.L.C. and colleagues thank the Rufford Foundation, Neotropical Grasslands Conservancy and CAPES for financing our study. We also thank Alexsander Zamorano Antunes for help in bird identification. Instituto Alexander con Humboldt, Reserva Natural La Planada, Smithsonian Institution. M.G.B. was supported by LTER8 NSF-DEB-2025755. Funding to M. Koivula came from the Academy of Finland, the Metsämiesten Säätiö Foundation, Metsäteho Ltd. and the Finnish Cultural Foundation. M.K.B. thanks the staff of DABCS, Legon; Environmental Protection Agency (EPA-Ghana), Plant Protection & Regulatory Services Directorate of the Ministry of Food & Agriculture (PPRSD-MoFA, Ghana). Meagan Grabowski was funded by the Yukon Fish and Wildlife Enhancement Trust, Yukon Parks and supported by Yukon University. Partial support for M. Goren's research was provided by the European Community's Seventh Framework Programme (FP7/2007-2013) for the project Vectors of Change in Oceans and Seas Marine Life, Impact on Economic Sectors (VECTORS) (B.S.G.). M.J.M. was supported by the Leverhulme Trust ECF-2021-512. MRW's research was facilitated by grants BSR-8811902, DEB-9411973, DEB-0080538, DEB-0218039, DEB-0620910, DEB-1239764, DEB-1546686 and DEB-1831952 from the National Science Foundation to the Institute of Tropical Ecosystem Studies, University of Puerto Rico and the International Institute of Tropical Forestry as part of the Long-Term Ecological Research Program. Additional support was provided by the United States Forest Service and the University of Puerto Rico. Although too numerous to recognise individually, a small army of undergraduates, a large number of graduate students and many post-doctoral associates have assisted with field collections over the past three decades. M.R.L. thanks the Instituto Felinos do Aguaí. N.A. thanks Karnataka Forest Department and IFP staff Messrs. S. Aravajy, S. Ramalingam, N. Barathan, G. Orukaimani, G. Jayapalan, K. Anthapa Gowda, Obbaya Gowda and Manoj Gowda. Funding for N.M.I. came from grant 99/09635-0, São Paulo Research Foundation (FAPESP). N. Beenaerts would like to thank Marc Janssens and students for collecting and identifying species. M. Milchakova acknowledges the IBSS State research assignments no.: 1023032000049-6-1.6.21. N. Valdivia was financially supported by the FONDECYT grant no.: 1230286 and FONDAP grant no.: 15150003 (IDEAL). N.A.J.G. thanks the Royal Society, Seychelles Fishing Authority, Seychelles Parks and Gardens Authority, Nature Seychelles. N. Poulet and colleagues wish to pay tribute to the numerous engineers and technicians who perform electrofishing and maintain the ASPE database. The database engineering and administration are funded by the Office Français de la Biodiversité (OFB, https://www.ofb. gouv.fr/). Nivaldo Peroni thanks CNPq for the productivity scholarship. O.F.L. would like to thank the Israeli Nature and Park Authority and all present and past Belmaker laboratory members who participated in data collection. O. Pizarro thanks the Australian Research Council, staff of Lizard Island Research Station. P. Hidalgo thanks ANID-Fondecyt 1191343 and regional Node ESPOBIS. PMvB and colleagues thank Operation Wallacea Ltd., which has supported long-term biodiversity surveys. We are grateful to the several thousand volunteers and scientists, guides, cooks and support staff, whose assistance has been invaluable during this period. We thank Expediciones y Servicios Ambientales Cusuco for providing logistical support and the Instituto de Conservación Forestal for issuing annual research permits allowing research in the Cusuco National Park to be completed. P.C.E. and colleagues thank A.S.B. Gontijo, D. Gontijo, I.S. Mendes, M.J. Correa, H. Kiefer, B. Pardinho, C. Araújo, I.M. Martins, V.S. Borges, J. Espanha, E. Quintana, J. Freitas, R. Viana, C. Moretzsohn and I.S. Barros for help during field work. P.S.P. was supported by the CNPq (302328/2022-0). P.A.-M. was supported by Funding National Doctoral Scholarship ANID (National Research and Development Agency), Chile.; no.: IC-120019 (IMO). Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for granting a research productivity scholarship to P. Higuchi. P.B.R. was supported by US NSF-DBI-2021898. P. Haase received funding from the EU Horizon 2020 project eLTER PLUS (grand agreement no.: 871128) and the DFG (German Research Foundation) project CRC 1439 RESIST (SFB 1439/1 2021–426547801). P.L.M. was supported by US National Science Foundation (LTREB), US AID Program, FONDECYT Chile and Northern Illinois University. P. Pysek thanks the long-term research development project RVO 67985939 (Czech Academy of Sciences). P.F.F. was supported by the Canada Graduate Scholarship (CGS D) and Michael Smith Foreign Study Supplement from the Natural Sciences and Engineering Research Council of Canada (NSERC). P. Valade acknowledges the DEAL Réunion, DEAL Mayotte, Office de l'eau de La Réunion. R.H.S. acknowledges all Bulgarian ornithologists and volunteers who collect data for birds. São Paulo forest plot data collection by RAFdL was supported by grant no.: 99/09635-0, São Paulo Research Foundation (FAPESP). For supporting the collection of the French Polynesian dataset, R. Beldade and colleagues would like to thank staff from the Centre de Recherches Insulaires et Observatoire de l'Environnement (CRIOBE). Agence National de la Recherche to R.B (ANR-14-CE02-0005-01/Stay or Go); Millenium Nucleus for the Ecology and Conservation of Temperate Mesophotic Reef Ecosystem (NUTME), Chile to R.B. RRR was funded by the Sao Paulo Research Foundation (Fapesp) grants 2013/50718-5 and 2020/05533-0. Fieldwork for R. Rocha was funded by a National Geographic Society grant EC-64368R-20. R.T.H. was funded by the (US) National Science Foundation. R.A.D. and colleagues are grateful to the Botanic Gardens and Parks Authority for ongoing support and previous funding for this project. Calum Irvine and Marcus Cosentino have assisted with data collections. The data from R.B.W. were upon work supported by the National Science Foundation under grants BSR-8811902, DEB-9411973, DEB-9705814, DEB-0080538, DEB-0218039, DEB-0620910, DEB-1239764, DEB-1546686 and DEB-1831952. Any opinions, findings, conclusions or recommendations expressed in the material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. We acknowledge the contribution of Jamarys Torres, International Institute of Tropical Forestry, USDA Forest Service, for collection of field data. R.D.H. acknowledges the Native Peoples of the North Slope of Alaska and the US National Science Foundation. Funding to R.J.M. was provided by the US National Science Foundation in support of the Santa Barbara Coastal Long-Term Ecological Research Program (award no.: OCE-1831937). The work by R.L.S. was supported by funding from the National Science Foundation to New Mexico State University for the Jornada Basin Long-Term Ecological Research Program (DEB 2025166). The study by R. Bottarin was co-funded by the European Union—NextGenerationEU, in the framework of the consortium iNEST—Interconnected Nord-Est Innovation Ecosystem (PNRR, Missione 4 Componente 2, Investimento 1.5 D.D. 1058 23/06/2022, ECS_00000043—Spoke1, RT3, CUP I43C22000250006) and the Autonomous Province of Bolzano. R. Pizzolotto would like to express sincere gratitude to the Paneveggio Regional Park Administration for their financial support. R.B.G. was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) fellowship number: 307671/2021-6. RvK was supported by the German Science Foundation (FZT 118). Work by R. Kyerematen was supported by a DAAD scholarship. We thank the Forestry Commission of Ghana and the Boti Falls Management for giving us access to the Study sites for this research. We also thank Mr. Henry Davis for helping with the identification of the specimens. R. Escribano was supported by ANID Chile grant ICN12_019. R.V. was supported by a doctoral grant from Fundação para a Ciência e Tecnologia, Portugal (SFRH/BD/84030/2012). Data for study 354 were sourced from Australia's Integrated Marine Observing System (IMOS)—IMOS is enabled by the National Collaborative Research Infrastructure Strategy (NCRIS). It is operated by a consortium of institutions as an unincorporated joint venture, with the University of Tasmania as Lead Agent. Marine National Facility acknowledgment: We acknowledge the use of the CSIRO Marine National Facility (https://ror.org/01mae9353) in undertaking part of this research. R.Y. and colleagues would like to thank the Israel Nature and Park Authority and all present and past Belmaker laboratory members who participated in data collection. R. Nouioua thanks Diogo F. Ferreira and Ricardo Rocha. S.E.M. extends sincere thanks to those involved in collecting the data (especially George LaRoi, Roger Hnatiuk, John Stadt and Benoit Gendreau-Berthiaume) and assisting with the archiving (Ellen Bledsoe, Amelia Hesketh, Justine Karst, Jenna Loesberg and Laura Super). The Portal Project was supported by research grants awarded to SKM Ernest and E.P. White by the US National Science Foundation (NSF DEB-1929730), USDA National Institute of Food and Agriculture, Hatch Project FLA-WEC-005983 (Ernest) and FLA-WEC-005944 (White). S.E.I.J. and colleagues thank Operation Wallacea and the staff and volunteers who have facilitated long-term work in Cusuco National Park, Honduras. SSJT contributed data included in this manuscript. Funding came from the National Science Foundation through grant DEB-1354563 to A.H. Hurlbert and E.P. White and by the Gordon and Betty Moore Foundation's Data-Driven Discovery Initiative through grant GBMF4563 to E.P. White. Funding for S.C.E. came from the Canadian IPY programme. S.S.G. and colleagues thank the Channel Islands National Park Service and Kelp Forest Monitoring Program divers for conducting underwater visual surveys. S.W.C. was supported by the National Research Foundation of Korea (RS-2023-00272745). S. Pico was supported by the Universidad de Cádiz (UCA/R93REC/2019). S.A.B. gratefully acknowledges the support of the German Centre of Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig (funded by the German Research Foundation; FZT 118). Data collection by S.A.M. was funded by Northern Beaches Council, Sydney Australia. S.Z.C. was supported by Bolsa PQ 307135/2020-9. S.K.A. thanks the Director, KFRI for the institutional support and KFRI Plan Fund for funding. The studies by S.V.B. and colleagues were funded by the Helmholtz Centre for Environmental Research—UFZ. Stefan Stoll acknowledges the State Environmental Agency of Rhineland-Palatinate, Germany. Data from S. Earl are based upon work supported by the National Science Foundation under grant no.: DEB-2224662, Central Arizona-Phoenix Long-Term Ecological Research Program (CAP LTER). Data from S.M.R. were provided by the H.J. Andrews Experimental Forest and Long-Term Ecological Research (LTER) Program, administered cooperatively by Oregon State University, the USDA Forest Service Pacific Northwest Research Station and the Willamette National Forest. This material is based upon work supported by the National Science Foundation under the grant LTER8 DEB-2025755. S.K.S. is grateful to the Bangladesh Forest Department for supporting data collection. The surveys by T. Gavriel were made possible thanks to the funding from the Israel Nature and Park Authority. We wish to thank all those who helped with fieldwork and the Interuniversity Institute for Marine Sciences in Eilat (IUI). T. Horton and colleagues thank the early scientific contributors, including David Billett, Tony Rice, Michael Thurston, Olaf Pfannkuche, Monty Priede and others, for their vision. We also thank the many graduate students and colleagues that have contributed to the data collection and analysis at the PAP-SO, and the British Oceanographic Data Centre for hosting and making the data available. The Porcupine Abyssal Plain—Sustained Observatory of the Natural Environment Research Council (NERC, UK) is principally funded through the Climate Linked Atlantic Sector Science (CLASS) project supported by NERC National Capability funding (NE/R015953/1). T. Caruso acknowledges the Latvian National LTER projects. The Treswell Wood Integrated Population Monitoring Group acknowledges the late John Michael McMeeking, MBE usually known as John McMeeking who was founder of the group remaining active in it until his death in 2019 and the late Neil A Taylor, usually known as Neil Taylor, who was a long-standing very active member of the group until his death in 2012. T. A. Bueno thanks FAPESP for grant no.: 2020/05533-0, São Paulo Research Foundation (FAPESP), that made possible the carry-on of this research. T.A.O. acknowledges the Local Environmental Observer Network contributors; M. Brubaker, M. Brook and staff of the Alaska Native Tribal Health Consortium; The Pew Charitable Trusts through the Pew Fellows Program in Marine Conservation. T.E.M. and colleagues thank the Operation Wallacea science teams who helped source data for some datasets. TGdS thanks the National Council for Scientific and Technological Development (CNPq) for financial support and Coordination of Superior Level Staff Improvement (CAPES) for scholarships. Funding to U. Klinck came from the BDF-F programme Lower Saxony; vegetation assessments by Bernadett Lambertz. Public data by V. Evtimova was provided by the Executive Environment Agency (ExEA), of the Ministry of Environment and Water, Bulgaria. Original data have been provided/collected by Wetlands International, as well as by specialists and experts from ExEA, RIEW and NGOs within the monitoring of wintering waterbirds as a part of the Bulgarian National Monitoring System for the condition of biodiversity. V.H.R.-M. acknowledges the National Science Foundation (NSF) through the Florida Coastal Everglades Long-Term Ecological Research (FCE LTER) program (#DBI-0620409, #DEB-9910514). V.S.S. was supported by the São Paulo Research Foundation, FAPESP (grant/award nos.: 2017/12138- 8, 2018/23111- 6); LTER- Itanhaém project (grant/award no.: 2012/51511-2). W. Beiroz and colleagues thank Grupo Jari for logistic support, GIS, soil, rainfall and timber production data. We thank the Conselho Nacional de Pesquisa e Desenvolvimento (CNPQ) and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) for funding the project (Site Peld 23—403811/2012-0). W.B. thanks Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for the PhD scholarship (BEX 3711-14-15). C.A.H.F. would like to thank his UNESP students Alexsander Seleguini, Angelo Luiz Tadeu Ottati, Cid Tacaoca Muraishi, Fabiana Oikawa and Sérgio Roberto Rodrigues, who worked on the identification of the species over the years, all local Farm employees for making possible the collections throughout all these years, with special thanks to Moacir José Ruela and the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, process no.: 99/08312-2) for financial support. All research by W.A.G. and colleagues at the USDA Forest Service International Institute of Tropical Forestry is done in collaboration with the University of Puerto Rico. All data from W.R.F. were obtained through financial support from the US National Science Foundation. Y.R.S. thanks the FUNDECT; CNPq; UEMS. The dataset of aerial and waterhole census in the Hwange National Park, Zimbabwe, was provided by the Wildlife and Environment Zimbabwe—Matabeleland Branch. Plankton data from S.P. was collected with support from Healthy Waterways, Queensland ARC LPO88366. JD Everett's research was funded by Australian Research Council Discovery Grant DP0880078.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
A static stable release of BioTIME version 2.0 can be found in Zenodo (https://doi.org/10.5281/zenodo.10932823). Code used in data curation and standardisation can be found at github.com/bioTIMEHub/BioTIME. The R package BioTIMEr is available in CRAN and can be found at github.com/bioTIMEHub/BioTIMEr.




