sPlotOpen – An environmentally balanced, open-access, global dataset of vegetation plots
Francesco Maria Sabatini and Jonathan Lenoir contributed equally to this work.
Abstract
Motivation
Assessing biodiversity status and trends in plant communities is critical for understanding, quantifying and predicting the effects of global change on ecosystems. Vegetation plots record the occurrence or abundance of all plant species co-occurring within delimited local areas. This allows species absences to be inferred, information seldom provided by existing global plant datasets. Although many vegetation plots have been recorded, most are not available to the global research community. A recent initiative, called ‘sPlot’, compiled the first global vegetation plot database, and continues to grow and curate it. The sPlot database, however, is extremely unbalanced spatially and environmentally, and is not open-access. Here, we address both these issues by (a) resampling the vegetation plots using several environmental variables as sampling strata and (b) securing permission from data holders of 105 local-to-regional datasets to openly release data. We thus present sPlotOpen, the largest open-access dataset of vegetation plots ever released. sPlotOpen can be used to explore global diversity at the plant community level, as ground truth data in remote sensing applications, or as a baseline for biodiversity monitoring.
Main types of variable contained
Vegetation plots (n = 95,104) recording cover or abundance of naturally co-occurring vascular plant species within delimited areas. sPlotOpen contains three partially overlapping resampled datasets (c. 50,000 plots each), to be used as replicates in global analyses. Besides geographical location, date, plot size, biome, elevation, slope, aspect, vegetation type, naturalness, coverage of various vegetation layers, and source dataset, plot-level data also include community-weighted means and variances of 18 plant functional traits from the TRY Plant Trait Database.
Spatial location and grain
Global, 0.01–40,000 m².
Time period and grain
1888–2015, recording dates.
Major taxa and level of measurement
42,677 vascular plant taxa, plot-level records.
Software format
Three main matrices (.csv), relationally linked.
1 BACKGROUND & SUMMARY
Biodiversity is facing a global crisis. As many as 1 million species are currently threatened with extinction, the vast majority due to anthropogenic impacts such as land-use and climate change (IPBES, 2019; WWF, 2020). In addition, the rates of biodiversity homogenization and redistribution are accelerating (Fricke & Svenning, 2020; Lenoir et al., 2020; Staude et al., 2020). Biological assemblages are becoming progressively more similar to each other globally, as local and endemic species go extinct and are replaced by more widespread and competitive native or alien species (IPBES, 2019; Staude et al., 2020). Many terrestrial and marine species are also shifting their geographical distribution as a response to climate change (Lenoir et al., 2020). This has profound potential impacts on ecosystems and human health (Bonebrake et al., 2018; Pecl et al., 2017).
Plant communities are no exception to this biodiversity crisis (Cardinale et al., 2011; Lenoir et al., 2008; Staude et al., 2020). This is particularly worrying since terrestrial vegetation accounts for 80% (450 Gt C) of the living biomass on Earth (Bar-On et al., 2018). Given the central role of vegetation in ecosystem productivity, structure, stability and functioning (Cardinale et al., 2011), assessing biodiversity status and trends in plant communities is paramount for other kingdoms of life and human societies alike.
Monitoring trends in plant biodiversity requires adequate data across a range of spatio-temporal scales (Kühl et al., 2020; Pimm, 2021). Large independent collections of plant occurrence data do exist at the global or continental extent via the Botanical Information and Ecology Network (BIEN; Enquist et al., 2016), the Global Inventory of Floras and Traits (GIFT; Weigelt et al., 2020) or the Global Biodiversity Information Facility (GBIF; https://www.gbif.org/). However, these databases suffer from one or several of the following limitations: (a) imbalance towards tree species only; (b) lack of data on how individual plant species co-occur and interact locally to form plant communities; and (c) coarse spatial resolutions (e.g., one-degree grid cells), which preclude intersection with high resolution remote sensing data and the assessment of biodiversity trends at the plant community level (Boakes et al., 2010).
There is a long tradition among botanists and phytosociologists to record the cover or abundance of each plant species that occurs in a vegetation plot (here used as a synonym of ‘relevé’ or ‘quadrat’) of a given size (i.e., surface area) at a given time (e.g., Stebler & Schröter, 1892). Compared to presence-only data, vegetation-plot data present many advantages. As all visible plant species are recorded, plots contain information on which plant species do, and do not co-occur in the same locality at a given moment in time (Chytrý et al., 2016). This is important for testing hypotheses related to biotic interactions among plant species. Vegetation-plot data also provide crucial information on where and when a species was absent, therefore, improving predictions from current species distribution models (Phillips et al., 2009). Being spatially explicit, vegetation plots can be resurveyed through time to assess potential changes in plant species composition relative to a baseline (Perring et al., 2018; Staude et al., 2020; Steinbauer et al., 2018). As they normally contain information on the relative cover or abundance of each species, vegetation plots are also more appropriate for detecting biodiversity changes than data representing only the occurrence of individual species (Beck et al., 2018; Jandt et al., 2011).
Globally, however, vegetation-plot data are very fragmented, as they typically stem from a myriad of local research and survey projects (Bruelheide et al., 2019). These are fine-grained data (e.g., 1–10,000 m2) normally covering small spatial extents (e.g., 1–1,000 km2). With their disparate sampling protocols, standards and taxonomic resolutions, aggregating and harmonizing vegetation plot data proves extremely challenging (Bruelheide et al., 2018). It is not surprising, therefore, that these data are rarely used in global-scale research on the biodiversity of plant communities (Aubin et al., 2020; Franklin et al., 2017; Wiser, 2016).
The sPlot initiative tries to close this data gap. It consolidates numerous local to regional vegetation-plot datasets to create a harmonized and comprehensive global database of georeferenced terrestrial plant species assemblages (Bruelheide et al., 2019). Established in 2013, sPlot v3.0 currently contains more than 1.9 million vegetation plots, and is fully integrated with the TRY database (Kattge et al., 2020), from which it derives information on plant functional traits. The sPlot database is increasingly being used to study continental-to-global scale vegetation patterns (Cai et al., 2021; Testolin, Attorre, et al., 2021; Testolin, Carmona, et al., 2021), such as the relative contribution of regional versus local factors to the global patterns of fern richness (Weigand et al., 2020), the mechanisms underlying the spread and abundance of native versus invasive tree species (van der Sande et al., 2020), and worldwide trait–environment relationships in plant communities (Bruelheide et al., 2018).
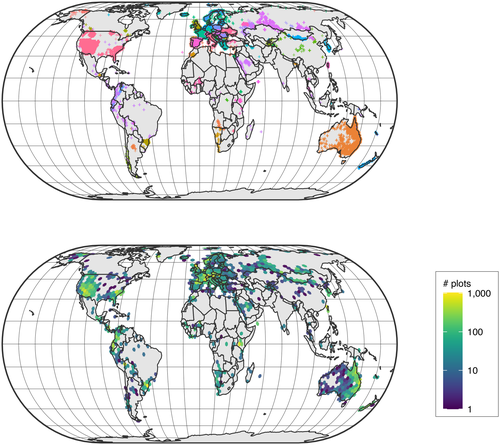
Yet, most of these data are not open-access. Here, we secured permission from data holders in the sPlot database to openly release a dataset composed of 95,104 vegetation plots. We selected the plots to be released using a replicated environmental stratification, in order to represent the entire environmental space covered by the sPlot database. This maximizes the benefits of releasing these data for a wide range of potential uses. The selected vegetation plots stem from 105 databases and span 114 countries (Figure 1). This resampled dataset (sPlotOpen – hereafter) is composed of: (a) plot-level information, including metadata and basic vegetation structure descriptors; (b) the vascular plant species composition of each vegetation plot, including species cover or abundance information when available; and (c) community-level functional information obtained by intersection with the TRY database (Kattge et al., 2020).
sPlotOpen is specifically designed for global macroecological studies, for example, the exploration of functional diversity patterns of communities with continental-to-global extent. We expect, however, that sPlotOpen might likewise prove useful to answer a range of different questions, related for instance to species co-occurrence patterns, the definition of species pools, the link between regional versus local determinants of species diversity, or the niche overlap between co-occurring species. Yet, data in sPlotOpen should not be considered as representative of the distribution of plant communities worldwide, especially when working at local spatial extents. This should be kept in mind for applications such as species distribution models (SDMs) or joint SDMs, whose results might be affected by the uneven geographical distribution of sPlotOpen's data. We refer the reader to the section ‘Usage notes’ for additional guidance on critical issues related, for instance, to incompletely sampled vegetation plots, varying plot size, and nested vegetation plots.
2 METHODS
2.1 Vegetation plot data sources
We started from the sPlot database v2.1 (created in October 2016), which contains 1,121,244 unique vegetation plots and 23,586,216 species records. Most of the data in sPlot refer to natural and semi-natural vegetation, while vegetation shaped by intensive and repeated human interference, such as cropland or ruderal communities, is hardly represented. Data originate from 110 different vegetation-plot datasets of regional, national or continental extent, some of which stem from regional or continental initiatives (see Bruelheide et al., 2019, for more information). For instance: 48 vegetation-plot datasets derive from the European Vegetation Archive (EVA; Chytrý et al., 2016); three major African datasets derive from the Tropical African Vegetation Archive (TAVA); and multiple vegetation datasets in the USA and Australia derive from the VegBank (Peet, Lee, Boyle, et al., 2012; Peet, Lee, Jennings, et al., 2012) and TERN’s AEKOS (Chabbi & Loescher, 2017) archives, respectively. Data from other continents (South America, Asia) or countries were contributed as separate standalone datasets. The metadata of each individual vegetation-plot dataset stored in sPlot are managed through the Global Index of Vegetation-Plot Databases (GIVD; Dengler et al., 2011), using the GIVD code as the unique dataset identifier.
2.2 Resampling method
Data in the sPlot database are unevenly distributed across vegetation types and geographical regions (see Bruelheide et al., 2018). Mid-latitude regions in developed countries (mostly Europe, the USA and Australia) are overrepresented in sPlot, while regions in the tropics and subtropics are underrepresented, which is a typical geographical bias in biodiversity data (see Lenoir et al., 2020; Lenoir & Svenning, 2015 for similar geographical bias in species redistribution). Such a geographical bias usually translates into an environmental bias with temperate climate usually more represented than tropical or Mediterranean climates. Unbalanced sampling effort in the environmental space is of particular concern for comparative macroecological studies (Bruelheide et al., 2018; Lenoir et al., 2010). To reduce this imbalance as much as possible, we performed a stratified resampling approach within the environmental space using several environmental variables available at global extent as sampling strata.
First, we removed vegetation plots without geographical coordinates or with a location uncertainty higher than 3 km. We also removed vegetation plots identified by the respective data contributors as having been recorded in wetlands or in anthropogenic vegetation types, since these data were available only for a few geographical regions, mostly in Europe. This resulted in a total of 799,400 out of the initial set of 1,121,244 vegetation plots.
We then ran a global principal component analysis (PCA) on a matrix of all terrestrial grid cells at a spatial resolution of 2.5 arcmin (n = 8,384,404), based on 30 climatic and soil variables. For climate, we used the 19 bioclimatic variables from CHELSA (Climatologies at high resolution for the earth's land surface areas) v1.2 (Karger et al., 2017), as well as two other bioclimatic variables reflecting the growing-season length (growing degree days above 1 ℃ – GDD1 – and 5 ℃ – GDD5), which were derived from CHELSA’s monthly temperatures as in Synes and Osborne (2011). In addition, we considered an index of aridity and a layer for potential evapotranspiration from the Consortium of Spatial Information (CGIAR-CSI, Trabucco & Zomer, 2010). For soil, we extracted seven variables from the SoilGrids database (Hengl et al., 2017), namely: (a) soil organic carbon content in the fine earth fraction; (b) cation exchange capacity; (c) pH; as well as the fractions of (d) coarse fragments; (e) sand; (f) silt; and (g) clay. The results of this PCA represent the full environmental space of all terrestrial habitats on Earth, irrespective of whether a grid cell hosted vegetation plots or not (Supporting Information Figure S1). We then subdivided the PCA ordination space, represented by the first two principal components (PC1–PC2), which accounted for 47 and 23%, respectively, of the total environmental variation in terrestrial grid cells, into a regular 100 × 100 grid. This PC1–PC2 two-dimensional space was subsequently used to balance our sampling effort across all PC1–PC2 grid cells for which vegetation plots were available. After excluding 42,878 vegetation plots for which no PC1 or PC2 values were available, due to missing data in the bioclimatic or soil variables, we projected the remaining 756,522 vegetation plots onto this PC1–PC2 grid. We finally calculated how many vegetation plots occurred in each PC1–PC2 grid cell (Figure 2).

In total, vegetation plots were available for 1,720 out of the 4,125 PC1–PC2 grid cells covered by the 8,384,404 terrestrial grid cells of the geographical space. We then resampled those PC1–PC2 grid cells (n = 858) with more than 50 vegetation plots, which is the median number of plots occurring across occupied grid cells in sPlot. This threshold of 50 vegetation plots represents a compromise between selecting a high number of plots, and keeping the resampled dataset as balanced as possible across the PC1–PC2 environmental space. To select these 50 vegetation plots we used the heterogeneity-constrained random resampling algorithm (Lengyel et al., 2011). This algorithm quantifies the variability in plant species composition among a set of vegetation plots by computing the mean and the variance of the Jaccard’s dissimilarity index (Jaccard, 1912) between all possible pairs of vegetation plots. More precisely, for a given PC1–PC2 grid cell containing more than 50 vegetation plots, we generated 1,000 random selections of 50 vegetation plots and ranked each selection according to the mean (ascending order) and variance (descending order) value of the Jaccard’s dissimilarity index. Ranks from both sortings were summed for each random selection, and the selection with the lowest summed rank was considered to provide the most balanced/even representation of vegetation types within the focal grid cell. Where a grid cell contained fewer than 50 plots, we retained all of them. In this way, we reduced the imbalance towards over-sampled climate types while ensuring that the resampled dataset represents the entire environmental gradient covered by the original sPlot database. This approach optimizes the selection of a subset of vegetation plots that encompasses the highest variability in species composition while avoiding peculiar and rare communities, which may represent outliers. As such, our approach maximizes variability over representativeness within each grid cell. We repeated the whole resampling procedure three times to get three different environmentally balanced, resampled subsets of our vegetation plots. These three resampling iterations can therefore be used as separate replicates, albeit these are not completely independent, as the same plots might have been drawn in two or even three of the three resampling iterations. In addition, those plots located in PC1–PC2 grid cells with fewer than 50 vegetation plots are completely shared by all three iterations.
2.3 Permission to release the data as open access
The resampling procedure resulted in 56,486, 56,501 and 56,494 vegetation plots selected during resampling iterations #1, #2 and #3, respectively, for a total of 107,238 unique vegetation plots. Since the sPlot database is a consortium of independent datasets whose copyright belongs to the data contributors, we used this preliminary potential selection to ask each dataset’s custodian (i.e., either the owner of a dataset or its authorized representative in the case of a collective dataset) for permission to release the data of selected vegetation plots as open access. For 12,134 unique vegetation plots, permission could not be granted because, for instance, the data are unpublished, confidential or sensitive. The number of vegetation plots for which the open-access permission was not granted in resampling iterations #1, #2 and #3 was 6,699, 6,690 and 6,705, respectively.
To mitigate the imbalance due to the exclusion of these confidential plots, we created a ‘consensus’ dataset. We started from resampling iteration #1, and replaced the 6,699 plots not granted as open access with plots selected in the second and third iterations, for which such permission could be granted (‘reserve’ plots, hereafter). We imposed the constraint that each candidate vegetation plot in the reserve pool should belong to the same environmental stratum, that is, the same PC1–PC2 grid cell, as the confidential vegetation plot, even though we acknowledge that this procedure does not maximize the variability in plant species composition of the replacement plots. Even after drawing from reserves, there were 3,150 plots that could not be replaced. These were distributed across 279 PC1–PC2 grid cells (16.2% of occupied cells), each cell having on average 11 irreplaceable plots (min. = 1, median = 5, max. = 50).
2.4 Trait information
For each vegetation plot for which open access could be granted, we computed the community-weighted mean and variance for 18 plant functional traits derived from the TRY database v3.0 (Kattge et al., 2020). These traits were selected among those that describe the leaf, wood, and seed economics spectra (Reich, 2014; Westoby, 1998), and are known to either affect different key ecosystem processes or respond to macroclimatic drivers, or both (Bruelheide et al., 2018). The 18 plant functional traits (all concentrations based on dry weight) were: (a) leaf area (mm2); (b) stem specific density (g/cm3); (c) specific leaf area (m2/kg); (d) leaf carbon concentration (mg/g); (e) leaf nitrogen concentration (mg/g); (f) leaf phosphorus concentration (mg/g); (g) plant height (m); (h) seed mass (mg); (i) seed length (mm); (j) leaf dry matter content (g/g); (k) leaf nitrogen per area (g/m2); (l) leaf N:P ratio (g/g); (m) leaf δ15N (per million); (n) seed number per reproductive unit; (o) leaf fresh mass (g); (p) stem conduit density (per mm2); (q) dispersal unit length (mm); and (r) conduit element length (μm).
Because missing values were particularly widespread in the species-trait matrix, we calculated community-weighted means using the gap-filled version of these traits we received from TRY (Kattge et al., 2020). Gap-filling was performed at the level of individual observations and relies on hierarchical Bayesian modelling (R package ‘BHPMF’ – Fazayeli et al., 2014; Schrodt et al., 2015) in R (R Core Team, 2020). This is a Bayesian machine learning approach, with no a priori assumptions, except for the data being missing completely at random. The algorithm ‘learns’ from the data, that is, if there was a phylogenetic signal in the data, this was used to fill the gaps but where no such signal was apparent, none was introduced. After gap-filling, we transformed to the natural logarithm all gap-filled trait values and averaged each trait by taxon (i.e., at species or genus level). The gap-filling approach was run only for species having at least one trait observation (n = 21,854). Additional information on the gap-filling procedure is available in Bruelheide et al. (2019).
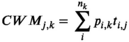 (1)
(1)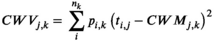 (2)
(2)3 DATA RECORDS
sPlotOpen contains 95,104 unique vegetation plots from 105 constitutive datasets (Table 1) and from 114 countries covering all continents except Antarctica (Figure 1). This is the result of pooling together the three environmentally balanced datasets from resampling iterations #1, #2 and #3 containing 49,787, 49,811 and 49,789 plots, respectively, after excluding the set of plots for which open access could not be granted by data contributors. The number of plots shared across all three resampling iterations is 19,672, while 14,939 plots are shared between two iterations. Replacing confidential plots in resampling iteration #1 with reserves from the other two iterations in the same PC1–PC2 grid cell resulted in a consensus version containing 53,262 plots. sPlotOpen only contains the species composition of vascular plants; information on the composition of bryophytes and lichens was discarded since it was only available for a minority of plots (n = 11,001 and n = 6,801, respectively). Information on the size (surface area) of the vegetation survey is available for 67,022 plots, and ranges between 0.03 and 40,000 m2 (mean = 377 m2; median = 100 m2). Specifically, sPlotOpen contains 12,894 plots with size smaller than 10 m2, 25,742 with size 10–100 m2, 24,750 plots with size 100–1,000 m2 and 3,075 plots with size greater or equal to 1,000 m2. Similarly, only for a minority of plots (n = 24,167) is information on the exact group of plants sampled in the field available (e.g., complete vegetation, only trees, only trees > 1 m height, and so on). However, as most data were collected using the phytosociological method, we deem it safe to assume that, unless otherwise specified, plots contain information on all vascular plants. We retained plots with incomplete vegetation, because they were mostly located in the tropics, that is, in areas where vegetation plots are particularly scarce otherwise. The average number of vascular plant species per vegetation plot ranges between 1 (i.e., monospecific stands) and 271 species (mean = 20; median = 16).
| GIVD ID | Dataset name | Custodian | Deputy custodian | No. open-access plots | Reference |
|---|---|---|---|---|---|
| 00-00-001 | ForestPlots.net | Oliver L. Phillips | Aurora Levesley | 169 | Lopez-Gonzalez et al. (2011) |
| 00-00-003 | SALVIAS | Brian Enquist | Brad Boyle | 3,403 | |
| 00-00-004 | Vegetation Database of Eurasian Tundra | Risto Virtanen | 519 | ||
| 00-00-005 | Tundra Vegetation Plots (TundraPlot) | Anne D. Bjorkman | Sarah Elmendorf | 309 | Elmendorf et al. (2012) |
| 00-RU-001 | Vegetation Database Forest of Southern Ural | Vasiliy Martynenko | Pavel Shirokikh | 68 | |
| 00-RU-002 | Database of Masaryk University’s Vegetation Research in Siberia | Milan Chytrý | 158 | Chytrý (2012) | |
| 00-RU-003 | Database Meadows and Steppes of Southern Ural | Sergey Yamalov | Mariya Lebedeva | 238 | |
| 00-TR-001 | Forest Vegetation Database of Turkey – FVDT | Ali Kavgacı | 45 | ||
| AF-00-001 | West African Vegetation Database | Marco Schmidt | Georg Zizka | 258 | Schmidt et al. (2012) |
| AF-00-003 | BIOTA Southern Africa Biodiversity Observatories Vegetation Database | Norbert Jürgens | Ute Schmiedel | 1,015 | Muche et al. (2012) |
| AF-00-006 | SWEA-Dataveg | Miguel Alvarez | Michael Curran | 1,675 | Alvarez et al. (2021) |
| AF-00-008 | PANAF Vegetation Database | Hjalmar S. Kühl | TeneKwetche Sop | 884 | |
| AF-00-009 | Vegetation Database of the Okavango Basin | Rasmus Revermann | Manfred Finckh | 378 | Revermann et al. (2016) |
| AF-BF-001 | Sahel Vegetation Database | Jonas V. Müller | Marco Schmidt | 556 | Müller (2003) |
| AF-CD-001 | Forest Database of Central Congo Basin | Kim Sarah Jacobsen | Hans Verbeeck | 140 | Kearsley et al. (2013) |
| AF-ET-001 | Vegetation Database of Ethiopia | Desalegn Wana | Anke Jentsch | 67 | Wana & Beierkuhnlein (2011) |
| AF-MA-001 | Vegetation Database of Southern Morocco | Manfred Finckh | 621 | Finckh (2012) | |
| AF-ZW-001 | Vegetation Database of Zimbabwe | Cyrus Samimi | 31 | Samimi (2003) | |
| AS-00-001 | Korean Forest Database | Tomáš Černý | Jiri Dolezal | 1,039 | Černý et al. (2015) |
| AS-00-003 | Vegetation of Middle Asia | Arkadiusz Nowak | Marcin Nobis | 314 | Nowak et al. (2017) |
| AS-00-004 | Rice Field Vegetation Database | Arkadiusz Nowak | 32 | ||
| AS-BD-001 | Tropical Forest Dataset of Bangladesh | Mohammed A. S. Arfin Khan | Fahmida Sultana | 87 | |
| AS-CN-001 | China Forest-Steppe Ecotone Database | Hongyan Liu | Fengjun Zhao | 117 | Liu et al. (2000) |
| AS-CN-002 | Tibet-PaDeMoS Grazing Transect | Karsten Wesche | Yun Jäschke | 58 | Wang et al. (2017) |
| AS-CN-003 | Vegetation Database of the BEF China Project | Helge Bruelheide | 24 | Bruelheide et al. (2011) | |
| AS-CN-004 | Vegetation Database of the Northern Mountains in China | Zhiyao Tang | 124 | ||
| AS-EG-001 | Vegetation Database of Sinai in Egypt | Mohamed Z. Hatim | 143 | Hatim (2012) | |
| AS-ID-001 | Sulawesi Vegetation Database | Michael Kessler | 24 | ||
| AS-IR-001 | Vegetation Database of Iran | Jalil Noroozi | Parastoo Mahdavi | 277 | |
| AS-KZ-001 | Database of Meadow Vegetation in the NW Tien Shan Mountains | Viktoria Wagner | 13 | Wagner (2009) | |
| AS-MN-001 | Southern Gobi Protected Areas Database | Henrik von Wehrden | Karsten Wesche | 1,032 | von Wehrden et al. (2009) |
| AS-RU-001 | Wetland Vegetation Database of Baikal Siberia (WETBS) | Victor Chepinoga | 9 | Chepinoga (2012) | |
| AS-RU-002 | Database of Siberian Vegetation (DSV) | Andrey Korolyuk | Andrei Zverev | 3,634 | Korolyuk & Zverev (2012) |
| AS-RU-004 | Database of the University of Münster – Biodiversity and Ecosystem Research Group's Vegetation Research in Western Siberia and Kazakhstan | Norbert Hölzel | Wanja Mathar | 207 | |
| AS-SA-001 | Vegetation Database of Saudi Arabia | Mohamed Abd El-Rouf Mousa El-Sheikh | 711 | El-Sheikh et al. (2017) | |
| AS-TJ-001 | Eastern Pamirs | Kim André Vanselow | 221 | Vanselow (2016) | |
| AS-TW-001 | National Vegetation Database of Taiwan | Ching-Feng Li | Chang-Fu Hsieh | 912 | |
| AS-YE-001 | Socotra Vegetation Database | Michele De Sanctis | Fabio Attorre | 236 | De Sanctis & Attorre (2012) |
| AU-AU-002 | AEKOS | Ben Sparrow | 10,976 | Chabbi & Loescher (2017) | |
| AU-NC-001 | New Caledonian Plant Inventory and Permanent Plot Network (NC-PIPPN) | Jérôme Munzinger | Philippe Birnbaum | 98 | Ibanez et al. (2014) |
| AU-NZ-001 | New Zealand National Vegetation Databank | Susan K. Wiser | 1,127 | Wiser et al. (2001) | |
| AU-PG-001 | Forest Plots from Papua New Guinea | Timothy J. S. Whitfeld | George D. Weiblen | 60 | Whitfeld et al. (2014) |
| EU-00-002 | Nordic-Baltic Grassland Vegetation Database (NBGVD) | Jürgen Dengler | Łukasz Kozub | 54 | Dengler & Rūsiņa (2012) |
| EU-00-011 | Vegetation-Plot Database of the University of the Basque Country (BIOVEG) | Idoia Biurrun | Itziar García-Mijangos | 2,142 | Biurrun et al. (2012) |
| EU-00-013 | Balkan Dry Grasslands Database | Kiril Vassilev | Armin Macanović | 269 | Vassilev et al. (2012) |
| EU-00-016 | Mediterranean Ammophiletea Database | Corrado Marcenò | Borja Jiménez-Alfaro | 783 | Marcenò & Jiménez-Alfaro (2017) |
| EU-00-017 | European Coastal Vegetation Database | John A. M. Janssen | 356 | ||
| EU-00-018 | The Nordic Vegetation Database | Jonathan Lenoir | Jens-Christian Svenning | 1,735 | Lenoir et al. (2013) |
| EU-00-019 | Balkan Vegetation Database | Kiril Vassilev | Hristo Pedashenko | 484 | Vassilev et al. (2016) |
| EU-00-020 | WetVegEurope | Flavia Landucci | 127 | Landucci et al. (2015) | |
| EU-00-022 | European Mire Vegetation Database | Tomáš Peterka | Martin Jiroušek | 2,560 | Peterka et al. (2015) |
| EU-AL-001 | Vegetation Database of Albania | Michele De Sanctis | Giuliano Fanelli | 31 | De Sanctis et al. (2017) |
| EU-AT-001 | Austrian Vegetation Database | Wolfgang Willner | Christian Berg | 2,310 | Willner et al. (2012) |
| EU-BE-002 | INBOVEG | Els De Bie | 119 | ||
| EU-BG-001 | Bulgarian Vegetation Database | Iva Apostolova | Desislava Sopotlieva | 160 | Apostolova et al. (2012) |
| EU-CH-005 | Swiss Forest Vegetation Database | Thomas Wohlgemuth | 2,134 | Wohlgemuth (2012) | |
| EU-CZ-001 | Czech National Phytosociological Database | Milan Chytrý | Ilona Knollová | 1,287 | Chytrý & Rafajová (2003) |
| EU-DE-001 | VegMV | Florian Jansen | Christian Berg | 15 | Jansen et al. (2012) |
| EU-DE-013 | VegetWeb Germany | Florian Jansen | Jörg Ewald | 587 | Ewald et al. (2012) |
| EU-DE-014 | German Vegetation Reference Database (GVRD) | Ute Jandt | Helge Bruelheide | 762 | Jandt & Bruelheide (2012) |
| EU-DK-002 | National Vegetation Database of Denmark | Jesper Erenskjold Moeslund | Rasmus Ejrnæs | 332 | |
| EU-ES-001 | Iberian and Macaronesian Vegetation Information System (SIVIM) – Wetlands | Aaron Pérez-Haase | Xavier Font | 580 | |
| EU-FR-003 | SOPHY | Emmanuel Garbolino | Patrice De Ruffray | 7,986 | Garbolino et al. (2012) |
| EU-GB-001 | UK National Vegetation Classification Database | John S. Rodwell | 3,182 | ||
| EU-GR-001 | KRITI | Erwin Bergmeier | 22 | ||
| EU-GR-005 | Hellenic Natura 2000 Vegetation Database (HelNatVeg) | Panayotis Dimopoulos | Ioannis Tsiripidis | 620 | Dimopoulos & Tsiripidis (2012) |
| EU-GR-006 | Hellenic Woodland Database | Ioannis Tsiripidis | Georgios Fotiadis | 17 | Fotiadis et al. (2012) |
| EU-HR-001 | Phytosociological Database of Non-Forest Vegetation in Croatia | Zvjezdana Stančić | 193 | Stančić (2012) | |
| EU-HR-002 | Croatian Vegetation Database | Željko Škvorc | Daniel Krstonošić | 585 | |
| EU-HU-003 | CoenoDat Hungarian Phytosociological Database | János Csiky | Zoltán Botta-Dukát | 46 | Lájer et al. (2008) |
| EU-IT-001 | VegItaly | Roberto Venanzoni | Flavia Landucci | 754 | Landucci et al. (2012) |
| EU-IT-010 | Vegetation database of Habitats in the Italian Alps – HabItAlp | Laura Casella | Pierangela Angelini | 247 | Casella et al. (2012) |
| EU-IT-011 | Vegetation-Plot Database Sapienza University of Rome (VPD-Sapienza) | Emiliano Agrillo | Fabio Attorre | 967 | Agrillo et al. (2017) |
| EU-LT−001 | Lithuanian Vegetation Database | Valerijus Rašomavičius | Domas Uogintas | 81 | |
| EU-LV-001 | Semi-natural Grassland Vegetation Database of Latvia | Solvita Rūsiņa | 369 | Rūsiņa (2012) | |
| EU-MK-001 | Vegetation Database of the Republic of Macedonia | Renata Ćušterevska | 28 | ||
| EU-NL-001 | Dutch National Vegetation Database | Stephan M. Hennekens | Joop H. J. Schaminée | 1,098 | Schaminée et al. (2006) |
| EU-PL-001 | Polish Vegetation Database | Zygmunt Kącki | Grzegorz Swacha | 692 | Kącki & Śliwiński (2012) |
| EU-RO-007 | Romanian Forest Database | Adrian Indreica | Pavel Dan Turtureanu | 166 | Indreica et al. (2017) |
| EU-RO-008 | Romanian Grassland Database | Eszter Ruprecht | Kiril Vassilev | 82 | Vassilev et al. (2018) |
| EU-RS-002 | Vegetation Database Grassland Vegetation of Serbia | Svetlana Aćić | Zora Dajić Stevanović | 217 | Aćić et al. (2012) |
| EU-RU-002 | Lower Volga Valley Phytosociological Database | Valentin Golub | Andrey Chuvashov | 383 | Golub et al. (2012) |
| EU-RU-003 | Vegetation Database of the Volga and the Ural Rivers Basins | Tatiana Lysenko | 174 | Lysenko et al. (2012) | |
| EU-RU-011 | Vegetation Database of Tatarstan | Vadim Prokhorov | Maria Kozhevnikova | 206 | Prokhorov et al. (2017) |
| EU-SI-001 | Vegetation Database of Slovenia | Urban Šilc | Filip Küzmič | 1,029 | Šilc (2012) |
| EU-SK-001 | Slovak Vegetation Database | Milan Valachovič | Jozef Šibík | 2,394 | Šibík (2012) |
| EU-UA-001 | Ukrainian Grasslands Database | Anna Kuzemko | Yulia Vashenyak | 301 | Kuzemko (2012) |
| EU-UA-006 | Vegetation Database of Ukraine and Adjacent Parts of Russia | Viktor Onyshchenko | Vitaliy Kolomiychuk | 96 | |
| NA-00-002 | Tree Biodiversity Network (BIOTREE-NET) | Luis Cayuela | 241 | Cayuela et al. (2012) | |
| NA-CA-003 | Database of Timberline Vegetation in NW North America | Viktoria Wagner | Toby Spribille | 63 | Wagner et al. (2014) |
| NA-CA-004 | Understory of Sugar Maple Dominated Stands in Quebec and Ontario (Canada) | Isabelle Aubin | 13 | Aubin et al. (2007) | |
| NA-CA-005 | Boreal Forest of Canada | Philippe Marchand | Yves Bergeron | 57 | Harper et al. (2003) |
| NA-GL-001 | Vegetation Database of Greenland | Birgit Jedrzejek | Fred J. A. Daniëls | 441 | Sieg et al. (2006) |
| NA-US-002 | VegBank | Robert K. Peet | Michael T. Lee | 14,965 | Peet, Lee, Jennings, et al. (2012) |
| NA-US-006 | Carolina Vegetation Survey Database | Robert K. Peet | Michael T. Lee | 3,263 | Peet, Lee, Boyle, et al. (2012) |
| NA-US-014 | Alaska-Arctic Vegetation Archive | Donald A. Walker | Amy Breen | 771 | Walker et al. (2016) |
| SA-00-002 | VegPáramo | Gwendolyn Peyre | Xavier Font | 2,010 | Peyre et al. (2015) |
| SA-AR-002 | Vegetation Database of Central Argentina | Melisa Giorgis | Alicia T. R. Acosta | 86 | |
| SA-BO-003 | Bolivia Forest Plots | Michael Kessler | Sebastian Herzog | 44 | |
| SA-BR-002 | Forest Inventory, State of Santa Catarina, Brazil (IFFSC Project) | Alexander Christian Vibrans | André Luís de Gasper | 1,561 | Vibrans et al. (2020) |
| SA-BR−003 | Grasslands of Rio Grande do Sul, Brazil | Eduardo Vélez-Martin | Valério D. Pillar | 306 | |
| SA-BR−004 | Grassland Database of Campos Sulinos | Gerhard E. Overbeck | Valério D. Pillar | 147 | |
| SA-CL−002 | SSAForests_Plots_db | Alvaro G. Gutiérrez | 155 | ||
| SA-CL-003 | Chilean Park Transects – Fondecyt 1040528 | Aníbal Pauchard | Alicia Marticorena | 44 | Pauchard et al. (2013) |
| SA-EC-001 | Ecuador Forest Plot Database | Jürgen Homeier | 166 |
Note
- Datasets are ordered based on their ID in the Global Index of Vegetation Databases (GVID ID).
By capping the number of vegetation plots in overrepresented environmental conditions, the resampling procedure described above strongly reduced the bias in the distribution of vegetation plots within the PC1–PC2 environmental space. Yet, due to the lack or scarcity of data from some geographical regions, like the tropics, there is some remaining imbalance in the spatial distribution of vegetation plots across geographical regions (Figure 1). This is evident when comparing the number of plots across continents. When considering the first resampling iteration only (n = 49,787), Europe is by far the best represented continent, with 15,920 vegetation plots. The least represented continents are Africa and South America, with 3,709 and 5,498 vegetation plots, respectively. Some residual imbalance remains also when considering biomes (Figure 3). With the exception of the ‘Temperate mid-latitudes’ biome, which includes 14,100 vegetation plots, all other biomes have a number of plots comprised between 1,558 (‘Polar and subpolar zone’) and 6,245 (‘Subtropics with year-round rain’) vegetation plots (Figure 3, left). Despite this residual imbalance, all the Whittaker biomes are covered by sPlotOpen (Figure 3, right), and our resampling algorithm has resulted in a much more balanced dataset than many other global datasets that are available, such as GBIF.

Almost one third of the 95,104 vegetation plots in sPlotOpen belong to forests (n = 38,282), one half to non-forest vegetation (n = 45,735), with 11.6% of plots remaining unassigned (n = 11,087). When not directly done by data providers, the assignment of plots to forests and non-forests was based on multiple lines of evidence, including the plot-level information on the cover of the tree layer, as well as traits of species composing a plot, such as growth form and height. In short, a plot record was considered as forest if the cover of the tree layer, or alternatively, the sum of the (relative) cover of all tree taxa (scaled by the sum of all cover values, as a percentage), was greater than 25%. It was considered a non-forest record if the sum of relative cover of low-stature, non-tree and non-shrub taxa was greater than 90%. For an extensive explanation of this classification scheme, we refer the reader to Bruelheide et al. (2019). Even though the proportion of forest versus non-forest vegetation plots is relatively well balanced, the geographical distribution of vegetation plots belonging to different vegetation types is likely not balanced in the geographical space, as it depends on the idiosyncrasies of the constitutive datasets composing the sPlot database. For instance, the data from New Zealand only include plots collected in non-forest ecosystems, while data from Chile only refer to forests. We urge potential users to carefully read the section ‘Usage notes’ below and the description of each individual dataset in GIVD (Dengler et al., 2011), and to contact the custodians of each dataset for further information.
4 DATABASE ORGANIZATION
The environmentally balanced and open-access dataset sPlotOpen is organized into three main matrices, relationally linked through the key column ‘PlotObservationID’.
The ‘header’ matrix contains plot-level information for the 95,104 vegetation plots, including: metadata (e.g., plot ID, data source, sampling date, geographical location, positional accuracy); sampling design information (e.g., the total surface area used during the vegetation survey); and a plot-level description of vegetation structure (e.g., vegetation type, percentage cover of each vegetation layer), vegetation type, and naturalness level (i.e., whether a plot belongs to the same formation that would occupy the site without human interference). Plots in Europe are also classified according to the European Nature Information System (EUNIS) habitat classification (column ‘ESY’), based on the habitat classification expert system (ESY, Chytrý et al., 2020). For each vegetation plot, we further provide information on the dataset it originates from, based on the IDs used in GIVD (Dengler et al., 2011). We also report four binary fields describing whether a plot belongs to the three resampling iterations (columns ‘Resample_1’, ‘Resample_2’, ‘Resample_3’), or to the first resampling iteration after the inclusion of replacement plots (column ‘Resample_1_consensus’). A brief summary of all the 47 variables in the header matrix is provided in Table 2.
| Variable | Range/levels | Unit of measurement | No. of plots with information | Type |
|---|---|---|---|---|
| GIVD_ID | see Table 1 | 95,104 | n | |
| Dataset | see Table 1 | 95,104 | n | |
| Continent | Africa, Asia, Europe, North America, Oceania, South America | 95,104 | n | |
| Country | 95,104 | n | ||
| Biome | Alpine, Boreal zone, Dry midlatitudes, Dry tropics and subtropics, Polar and subpolar zone, Subtropics with year-round rain, Subtropics with winter rain, Temperate midlatitudes, Tropics with summer rain, Tropics with year-round rain | 95,104 | n | |
| Date_of_recording | 05-07-1888 - 03-02-2015 | dd-mm-yyyy | 80,085 | d |
| Latitude | −54.82303 – 80.149116 | ° (WGS84) | 95,104 | q |
| Longitude | −162.741433 – 176.4221 | ° (WGS84) | 95,104 | q |
| Location_uncertainty | 1–2,750 | m | 95,075 | q |
| Releve_area | 0.03–40,000 | m2 | 67,022 | q |
| Plant_recorded | All vascular plants, All trees & dominant understory, Dominant trees, Only dominant species, Dominant woody plants >= 2.5 cm dbh, All woody plants, Woody plants >= 1 cm dbh, Woody plants >= 2.5 cm dbh, Woody plants >= 5 cm dbh, Woody plants >= 10 cm dbh, Woody plants >= 20 cm dbh, Woody plants >= 1 m height, Not specified | 95,104 | n | |
| Elevation | −30 – 5,960 | m a.s.l. | 62,968 | q |
| Aspect | 1–360 | ° | 42,178 | q |
| Slope | 0–90 | ° | 51,246 | q |
| is_forest | FALSE = 45,735; TRUE = 38,282 | 84,017 | b | |
| ESY | 39,632 | n | ||
| Naturalness | 1 = Natural, 2 = Semi-natural | 60,192 | o | |
| Forest | FALSE = 36,282; TRUE = 33,170 | 69,452 | b | |
| Shrubland | FALSE = 58,245; TRUE = 11,207 | 69,452 | b | |
| Grassland | FALSE = 33,800; TRUE = 35,652 | 69,452 | b | |
| Wetland | FALSE = 59,196; TRUE = 10,256 | 69,452 | b | |
| Sparse_vegetation | FALSE = 66,177; TRUE = 3,275 | 69,452 | b | |
| Cover_total | 1–990 | % | 19,407 | q |
| Cover_tree_layer | 0.5–150 | % | 12,094 | q |
| Cover_shrub_layer | 0.5–170 | % | 16,804 | q |
| Cover_herb_layer | 0.2–199 | % | 29,668 | q |
| Cover_moss_layer | 1–100 | % | 9,681 | q |
| Cover_lichen_layer | 1–90 | % | 708 | q |
| Cover_algae_layer | 1–100 | % | 41 | q |
| Cover_litter_layer | 1–107 | % | 3,161 | q |
| Cover_bare_rocks | 1–100 | % | 2,747 | q |
| Cover_cryptogams | 1–90 | % | 772 | q |
| Cover_bare_soil | 0–99 | % | 2,746 | q |
| Height_trees_highest | 1–99 | m | 8,220 | q |
| Height_trees_lowest | 1–90 | m | 447 | q |
| Height_shrubs_highest | 0.1–9.9 | m | 3,389 | q |
| Height_shrubs_lowest | 0.1–9 | m | 263 | q |
| Height_herbs_average | 0.1–600 | cm | 5,901 | q |
| Height_herbs_lowest | 1–150 | cm | 490 | q |
| Height_herbs_highest | 1–600 | cm | 1,083 | q |
| SoilClim_PC1 | −6.233 – 8.172 | 95,104 | q | |
| SoilClim_PC2 | −4.824 – 15.466 | 95,104 | q | |
| Resample_1 | FALSE = 45,317; TRUE = 49,787 | 95,104 | b | |
| Resample_2 | FALSE = 45,293; TRUE = 49,811 | 95,104 | b | |
| Resample_3 | FALSE = 45,315; TRUE = 49,789 | 95,104 | b | |
| Resample_1_consensus | FALSE = 41,842; TRUE = 53,262 | 95,104 | b |
Note
- dbh = diameter at breast height. Variable types can be n = nominal (i.e., qualitative variable); o = ordinal; q = quantitative; b = binary (i.e., Boolean); or d = date. Additional details on the variables are in Bruelheide et al. (2019). Global Index of Vegetation Databases (GIVD) codes derive from Dengler et al. (2011). Biomes refer to Schultz (2005), modified to include also the world mountain regions (Körner et al., 2017). The column ESY refers to the European Nature Information System (EUNIS) Habitat Classification expert system (ESY, Chytrý et al., 2020).
The ‘DT’ matrix contains data on the species composition of each plot. It is structured in a long format and contains 1,945,384 records from 42,680 vascular plant taxa, mostly resolved at the species level. For each record, we report both the taxon name as originally contributed by the data custodian (column ‘Original_species’), and the taxon name after taxonomic standardization (column ‘Species’). For details on the taxonomic standardization, please see section ‘Technical validation’ below. For each species we also provided cover/abundance values. These follow different standards across the datasets constituting the sPlot database. We, therefore, provide both the cover/abundance value as reported in the original data (column ‘Original_abundance’), together with the abundance scale that was originally used (column ‘Abundance_scale’). This can take seven values: ‘CoverPerc’ = percentage cover; ‘pa’ = presence-absence; ‘x_BA’ = basal area (m2/ha, only for woody species); ‘x_IC’ = individual count, that is, number of individuals in plot; ‘x_SC’ = stem count, that is, number of stems in plot; ‘x_IV’ = importance value index; and ‘x_PF’ = presence frequency. The great majority of entries, however, use the percentage cover scale (n = 1,709,000). Finally, for each entry, we calculated a ‘Relative_cover’, that is, the cover/abundance of a given taxon divided by the total cover/abundance of all taxa in that vegetation plot.
The ‘CWM_CWV’ matrix contains the community-weighted means and variances calculated for each of the 18 functional traits mentioned above. It also contains three additional columns. The column ‘Species_richness’ shows the number of species recorded in each plot. The columns ‘Trait_coverage_cover’ and ‘Trait_coverage_pa’ provide, respectively, the proportion of total cover and the proportion of species in a plot for which functional trait information was available. In total, functional trait information was available for 21,854 species. As functional trait information was based on gap-filled data (see above), each of these 21,854 species had information for all the 18 functional traits. The average proportion of species in each plot for which functional trait information was available is .85 (median = .95). For 42,012 plots, the coverage was complete, while we do not have functional trait information for any of the species occurring in 482 plots. When considering relative cover, the average trait coverage is .87, with 74,151 plots having functional trait information for species cumulatively accounting for more than 80% of relative cover. When considering the number of species, 68,041 plots have functional trait information for 80% or more of the species occurring in that plot.
sPlotOpen contains two additional objects. The ‘metadata’ matrix contains plot-level metadata, which provide information on the origin of each individual vegetation plot. This object contains 15 columns, with information on the dataset of origin (column ‘GIVD_ID’ – Dengler et al., 2011), author or surveyor names (columns ‘Releve_author’ and ‘Releve_coauthor’), bibliographic references both at the dataset (column ‘DB_BIBTEXKEY’) and plot level (‘Plot_Biblioreference’ and ‘BIBTEXKEY’), when available. Similarly, the column ‘Project_name’ provides information on the project in which a vegetation plot was originally recorded. When available, we also provide information on the numbering of the plots in the publication where they originally appeared (columns ‘Nr_table_in_publ’, ‘Nr_releve_in_table’), or in the dataset where they were initially stored (‘Original_nr_in_database’). In the case of nested plots (n = 1,851), we also provide the original plot and subplot IDs (columns: ‘Original_plotID’, ‘Original_subplotID’). The last two columns report plot-level ‘Remarks’, and the unique identifier produced by Turboveg when the vegetation plot was first stored (‘GUID’). Turboveg is a program specifically designed to store, maintain and export vegetation plot data (https://www.synbiosys.alterra.nl/turboveg; Hennekens & Schaminée, 2001).
Finally, the object ‘references’ contains all the bibliographic references formatted according to a BibTex standard. Each reference is tagged with a key corresponding to the fields ‘DB_BIBTEXKEY’ and ‘BIBTEXKEY’ in the metadata. We further provide an R function (‘sPlotOpen_citation’) to create reference lists, based on a selection of plots and/or datasets.
Except for the ‘reference’ file (format.bib), all objects/matrices are provided in tab-delimited .txt files. All objects, including the ‘sPlotOpen_citation’ function, are also compiled inside a .RData object.
5 TECHNICAL VALIDATION
The original sPlot database has a nested structure and consists of several individual datasets, each validated and maintained by its respective dataset custodian. In many cases, individual datasets are also collections whose vegetation plots were provided by their respective owners (the person who performed the actual vegetation survey) or by someone who digitized the original data from the scientific published or grey literature. We obviously have no direct control over the individual vegetation plots that we provide here in sPlotOpen. Yet, all these vegetation plots stem from trained professional botanists, or published scientific work, and are accompanied by detailed information on the sampling protocols used, thus ensuring data quality and reliability.
Before integration into the sPlot database, each dataset was further checked for consistency. If the dataset was in a different format, we converted it to a Turboveg 2 dataset (Hennekens & Schaminée, 2001). During this conversion, we checked that all datasets contained the required metadata information, and cross-checked that each plot was located within the geographical scope of its respective dataset. All individual Turboveg 2 datasets were then integrated into a Turboveg 3 database, and exported to comma-separated files. Finally, we harmonized all the taxonomic names from all datasets, based on sPlot’s taxonomic backbone (Purschke, 2017). This backbone matched all the taxonomic names (without nomenclatural authors) from all datasets in sPlot v2.1 and TRY v3.0 (Kattge et al., 2020) to their resolved version based on the Taxonomic Name Resolution Service web application (TNRS v4.0; Boyle et al., 2013). This allowed us to (a) harmonize all datasets to a common nomenclature and (b) link the sPlot database to the TRY database (Kattge et al., 2020). The final backbone only retained matched taxonomic names at the rank of species or higher. Additional detail on the taxonomic resolution is reported in Bruelheide et al. (2019), while a description of the workflow, including R-code, is available in Purschke (2017).
6 USAGE NOTES
The sPlotOpen database can be downloaded from https://doi.org/10.25829/idiv.3474-40-3292. A short vignette introducing the use of sPlotOpen in R can be found in Supporting Information Appendix S1. Users are urged to cite the original sources when using sPlotOpen in addition to the present paper (see Table 1). For two datasets (AF-00–009, AF-CD-001), the identification of taxa at species level is still in progress. Data on lichens and mosses, where available (e.g., dataset NA-GL-001), can be obtained on request from the respective dataset custodian or sPlot coordinator. As most of the constitutive datasets remain under continuous development, sPlotOpen users are encouraged to get in touch with the custodian(s) of the data they are planning to use (the updated list of custodian names is maintained on the sPlot website).
The use of sPlotOpen comes with a number of warnings. First, sPlotOpen was resampled in a way that maximizes the compositional variability of vegetation in different environmental conditions. As such, sPlotOpen should not be considered as representative of the spatial distribution of plant communities, especially when the focus has a local or regional spatial extent. Second, for most regions, data were collected opportunistically, and without a randomized sampling design. This might lead to some vegetation types being oversampled in some regions, but undersampled in other regions, which might affect the output of species distribution models, especially at local or regional spatial extents. Third, not all plots were sampled using the same plot size, and some plots, mostly located in tropical regions, only contain data on woody species. This should be accounted for when exploring biodiversity patterns or comparing biodiversity indices (e.g., species richness, beta diversity) across plots or regions. Finally, a small fraction of plots are nested subsets of larger plots. Depending on the application, this might or might not represent a problem. Nested plots can be identified using the information in the ‘metadata’ matrix. The most appropriate way to deal with these issues depends on the problem being analysed. Users are, therefore, invited to carefully consider the limitations above when designing applications relying on sPlotOpen.
The data described here represent the subset of sPlot for which we were able to secure permission for making these data open. Additional data from sPlot are available under sPlot’s Governance and Data Property Rules (https://www.idiv.de/en/splot). Using the full sPlot dataset is also recommended if a stratification is desired that is different from the environmental factors used here, for example by geographical region or plot size.
ACKNOWLEDGMENTS
The authors are grateful to the thousands of vegetation scientists who sampled vegetation plots in the field or digitized them into regional, national or international databases. The authors also appreciate the support of the German Research Foundation for funding sPlot as one of the iDiv (DFG FZT 118, 202548816) research platforms, as well as for funding the position of Francesco Maria Sabatini and the organization of three workshops through the sDiv calls. The authors acknowledge this support with naming the database ‘sPlot’, where the ‘s’ refers to the sDiv synthesis workshops. The authors are also grateful to Anahita Kazem and iDiv's Data & Code Unit for assistance with curation and archiving of the dataset.
The study has been supported by the TRY initiative on plant traits (http://www.try-db.org). The TRY initiative and database is hosted, developed and maintained by J. Kattge and G. Bönisch (Max Planck Institute for Biogeochemistry, Jena, Germany). TRY is currently supported by DIVERSITAS/Future Earth and iDiv Halle-Jena-Leipzig. Jens Kattge acknowledges support by the Max Planck Institute for Biogeochemistry (Jena, Germany), Future Earth, iDiv Halle-Jena-Leipzig and the EU H2020 project BACI, Grant No. 640176.
Isabelle Aubin was funded through the Natural Sciences and Engineering Research Council of Canada and Ontario Ministry of Natural Resources and Forestry. Yves Bergeron was funded through the Natural Sciences and Engineering Research Council of Canada. Idoia Biurrun was funded by the Basque Government (IT936-16). Anne Bjorkman thanks the Herschel Island-Qikiqtaruk Territorial Park management, Catherine Kennedy, Dorothy Cooley, Jill F. Johnstone, Cameron Eckert and Richard Gordon for establishing the ecological monitoring programme. Funding was provided by Herschel Island-Qikiqtaruk Territorial Park. Luis Cayuela was supported by project BIOCON08_044 funded by Fundación BBVA (Banco Bilbao Vizcaya Argantiera). Milan Chytrý, Flavia Landucci, Corrado Marcenò and Tomáš Peterka were supported by the Czech Science Foundation (project no. 19-28491X). Brian Enquist thanks the following individuals and institutions for contributing data to sPlot via the SALVIAS database: Mauricio Bonifacino, Saara DeWalt, Timothy Killeen, Susan Letcher, Nigel Pitman, Cam Webb, The Missouri Botanical Garden, RAINFOR and the Amazon Forest Inventory Network. Alvaro G. Gutiérrez was funded by Project FORECOFUN-SSA PIEF-GA-2010–274798 and FONDECYT 1200468. Mohamed Z. Hatim thanks Kamal Shaltout and Joop Schaminée for MSc thesis supervision, and Joop Schaminée for support and funding from the Prince Bernard Culture Fund Prize for Nature Conservation. Jürgen Homeier received funding from BMBF (Federal Ministry of Education and Science of Germany) and the German Research Foundation (DFG Ho3296-2, DFG Ho3296-4). Borja Jiménez-Alfaro was funded by the Spanish Research Agency through grant AEI/10.13039/501100011033. Dirk N. Karger received funding from: the Swiss Federal Institute for Forest, Snow and Landscape Research (WSL) internal grant exCHELSA and ClimEx, the Joint Biodiversa COFUND project ‘FeedBaCks' and ‘Futureweb', the Swiss Data Science Projects: SPEEDMIND, and COMECO, and the Swiss National Science Foundation (20BD21_184131). Hjalmar Kühl gratefully acknowledges the Pan African team and funding by the Max Planck Society and Krekeler Foundation. Attila Lengyel was supported by the National Research, Development and Innovation Office, Hungary (PD-123997). Tatiana Lysenko was funded by the Russian Foundation for Basic Research (Grant No. 16-04-00747a). Alireza Naqinezhad is supported by a master grant from the University of Mazandaran. Jérôme Munzinger was supported by the French National Research Agency (ANR) with grants INC (ANR-07-BDIV-0008), BIONEOCAL (ANR-07-BDIV-0006) & ULTRABIO (ANR-07-BDIV-0010), by the National Geographic Society (Grant 7579-04), and with funding and authorizations of North and South Provinces of New Caledonia. Arkadiusz Nowak received support from the National Science Centre, Poland, grant no. 2017/25/B/NZ8/00572. Gerhard E. Overbeck acknowledges support from Brazil's National Council of Scientific and Technological Development (CNPq, grant 310022/2015-0). Meelis Pärtel was supported by the Estonian Research Council (PRG609) and European Regional Development Fund (Centre of Excellence EcolChange). Robert Peet acknowledges the support from the National Center for Ecological Analysis and Synthesis, the North Carolina Ecosystem Enhancement Program, the U.S. Forest Service, and the U.S. National Science Foundation (DBI-9905838, DBI-0213794). Josep Peñuelas acknowledges the financial support from the European Research Council Synergy grant ERC-SyG-2013-610028 IMBALANCE-P. Petr Petřík and Jiri Dolezal acknowledge the support of the long-term research development project No. RVO 67985939 of the Czech Academy of Sciences. Oliver Phillips was funded by an ERC Advanced Grant (291585, ‘T-FORCES’) and a Royal Society-Wolfson Research Merit Award. Valério D. Pillar was supported by the Brazil's National Council of Scientific and Technological Development (CNPq, grant 307689/2014-0). Solvita Rūsiņa was supported by the University of Latvia grant AAP2016/B041//Zd2016/AZ03 within the ‘Climate change and sustainable use of natural resources’ framework. Franziska Schrodt was supported by the University of Minnesota Institute on the Environment Discovery Grant, the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig grant (50170649_#7) and the University of Nottingham Anne McLaren Fellowship. Jozef Šibík was funded by The Slovak Research and Development Agency grant no. APVV16-0431. Jens-Christian Svenning considers this work a contribution to his VILLUM Investigator project ‘Biodiversity Dynamics in a Changing World’ funded by VILLUM FONDEN (grant 16549). Kim André Vanselow would like to thank W. Bernhard Dickoré for the help in the identification of plant species and acknowledges the financial support from the Volkswagen Foundation (AZ I/81 976) and the German Research Foundation (DFG VA 749/1-1, DFG VA 749/4-1). Evan Weiher was funded by NSF DEB-0415383, UWEC-ORSP, and UWEC-BCDT. Work by Karsten Wesche was supported by the German Research Foundation (DFG WE 2601/3-1,3-2, 4-1,4-2) and by the German Ministry for Science and Education (BMBF, CAME 03G0808A). Susan Wiser was funded by the New Zealand (NZ) Ministry for Business, Innovation and Employment's Strategic Science Investment Fund.
This paper is dedicated to the memory of Dr. Ching-Feng (Woody) Li.
CONFLICT OF INTEREST
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
FMS wrote the first draft of the manuscript, with considerable input from JL and HB. JL and TH wrote the resampling algorithm. FMS set up the GitHub projects, curated the database, and produced the graphs. He also coordinated the sPlot consortium. SMH wrote the Turboveg software, which holds the sPlot database. JKa provided the trait data from TRY and FSc performed the trait data gap filling. HB secured the funding for sPlot as a strategic project of iDiv. All other authors contributed data and/or helped set up the database and/or helped develop the resampling algorithm. All authors contributed to revising and approved the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The R code used to produce sPlotOpen from the sPlot v2.1 database is contained in the sPlotOpen_code GitHub repository: https://github.com/fmsabatini/sPlotOpen_Code. This manuscript was produced using the Manubot workflow (Himmelstein et al., 2019). The code for reproducing this manuscript is stored in the sPlotOpen_manuscript GitHub repository: https://github.com/fmsabatini/sPlotOpen_Manuscript.
REFERENCES
BIOSKETCH
sPlot is a collaborative initiative to integrate existing local and national vegetation-plot datasets into a global harmonized database. It was initiated in 2013, within the sDiv working group ‘Plant trait-environment relationships across the world’s biomes’. Since then, it has become established as the largest vegetation-plot database worldwide and coordinates a consortium of 251 individual active members, representing 167 local and national datasets. sPlot’s overarching scientific goal is the exploration of all aspects of global plant community diversity, including taxonomic, functional and phylogenetic diversity, across biomes, vegetation types, taxonomic or functional guilds and scales. Central to sPlot’s mission is the exploration of the relationships between environmental drivers, trait variation, and assembly processes in local plant communities worldwide.




