Distinct spread of DNA and RNA viruses among mammals amid prominent role of domestic species
Abstract
Aim
Emerging infectious diseases arising from pathogen spillover from mammals to humans constitute a substantial health threat. Tracing virus origin and predicting the most likely host species for future spillover events are major objectives in One Health disciplines.
We assessed patterns of virus sharing among a large diversity of mammals, including humans and domestic species.
Location
Global.
Time period
Current.
Major taxa studied
Mammals and associated viruses.
Methods
We used network centrality analysis and trait-based Bayesian hierarchical models to explore patterns of virus sharing among mammals. We analysed a global database that compiled the associations between 1,785 virus species and 725 mammalian host species as sourced from automatic screening of meta-data accompanying published nucleotide sequences between 1950 and 2019.
Results
We show that based on current evidence, domesticated mammals hold the most central positions in networks of known mammal–virus associations. Among entire host–virus networks, Carnivora and Chiroptera hold central positions for mainly sharing RNA viruses, whereas ungulates hold central positions for sharing both RNA and DNA viruses with other host species. We revealed strong evidence that DNA viruses were phylogenetically more host specific than RNA viruses. RNA viruses exhibited low functional host specificity despite an overall tendency to infect phylogenetically related species, signifying high potential to shift across hosts with different ecological niches. The frequencies of sharing viruses among hosts and the proportion of zoonotic viruses in hosts were larger for RNA than for DNA viruses.
Main conclusions
Acknowledging the role of domestic species in addition to host and virus traits in patterns of virus sharing is necessary to improve our understanding of virus spread and spillover in times of global change. Understanding multi-host virus-sharing pathways adds focus to curtail disease spread.
1 INTRODUCTION
Pathogen spillover and cross-species transmission between animals and humans is a major source of infectious diseases and a considerable global public health burden (Jones et al., 2008; Karesh et al., 2012). Understanding the factors that enable or facilitate these processes is a crucial step for such events to be predicted. Host shifting, that is the colonization of a new host species by a pathogen, requires a certain level of overlap in species traits (“ecological fitting”) in order to overcome barriers of cross-species transmission and for survival and reproduction within novel host species (Agosta, Janz, & Brooks, 2010; Parrish et al., 2008; Woolhouse, Haydon, & Antia, 2005). In the search for mechanisms and enabling conditions that might help to predict the future emergence of infectious diseases from animal populations, the necessity of considering entire host species communities amongst underpinning biogeographical structure and connectivity has recently been emphasized (Clark et al., 2018; Fenton, Streicker, Petchey, & Pedersen, 2015; Poulin, 2010; Wells et al., 2018).
Network analyses that describe the connections of different host species in terms of parasite sharing have proved useful in analysing host specificity and parasite spread (Gómez, Nunn, & Verdú, 2013; Luis et al., 2015), particularly given that they offer the opportunity to explore community-wide pathogen spread (the distribution of a pathogen among host species, a pattern emerging from past and contemporary host-shifting events that connect host species as nodes in a network). Other recent “big data” studies of mammal–virus associations have explored whether host traits and geographical distribution can predict those species that are most likely to harbour undiscovered viruses that might cause future pandemics using trait-based regression analysis (Han, Schmidt, Bowden, & Drake, 2015; Luis et al., 2015; Olival et al., 2017). Such approaches might lead to increased predictability of future pandemics.
Nevertheless, despite important advances in virus discovery and analytical approaches, our understanding of virus sharing and their spread through entire networks of mammalian host species remains limited. The challenge of assessing different animal species in their role for virus spread is understandable, because detailed information about virus sharing across entire communities has only become available recently (Olival et al., 2017; Wardeh, Risley, McIntyre, Setzkorn, & Baylis, 2015) amid the challenge that many virus species remain unknown (Carroll et al., 2018).
We address this knowledge gap by exploring the role of different mammalian species in the spread of viruses through entire host communities. In particular, we test whether domestic species (livestock and companion animals) play a major role in virus spread and spillover among humans and wildlife. To this end, there are strong reasons why domesticated animals should cover central positions in networks of host–virus associations. Domesticated animals share large numbers of viruses and other parasites with humans (Morand, McIntyre, & Baylis, 2014) and were recently reported to play crucial roles in the sharing of helminth parasites between humans and wildlife (Wells et al., 2018). Moreover, the large numbers of domestic animals compared with the numbers of wildlife (Bar-On, Phillips, & Milo, 2018), and the close contact between them and people, creates ground for frequent and multilateral exposure. For entire networks of viruses and mammalian host associations, we also expect different patterns of virus sharing for the two different genome types of DNA and RNA viruses. Greater rates of replication error and higher genetic diversity in RNA virus populations have been proposed to increase their host range through more frequent host shifting and adaptation to distantly related host species, whereas DNA viruses and retroviruses are assumed to be more host specific owing to stronger co-divergence with their hosts over much longer evolutionary time-scales (Cleaveland, Laurenson, & Taylor, 2001; Geoghegan, Duchêne, & Holmes, 2017; Jackson & Charleston, 2004; Longdon et al., 2018). With the mounting recognition that host use in parasites seems to be more constrained by ecological opportunity than by evolutionary history, there is an urgent need to understand and quantify pathogen spread and host-shifting capacity in response to specific traits at a global scale (Nylin et al., 2018; Wells & Clark, 2019). Nevertheless, to date little comprehensive work has explored whether host sharing and virus spread at the network level differ among these types of viruses and whether they interact with the various groups of mammals in different ways. We used network centrality analysis and Bayesian hierarchical models to quantify the extent of virus sharing among different mammalian host species and the proportion of zoonotic viruses carried in different hosts. If domestic species are key drivers of virus spread, we expect them to occupy central positions in networks of pathogen sharing at the human–domestic animal–wildlife interface, whereby variation in the host specificity of viruses might curtail their spread among the diversity of mammalian hosts at the global scale.
2 METHODS
2.1 Virus–host data
We extracted mammal–virus species-level interactions from the Enhanced Infectious Diseases Database (EID2) (Wardeh et al., 2015) in the version from March 2019. In brief, EID2 uses automated mining procedures to extract information on pathogens, their hosts and locations from two sources: (a) the meta-data accompanying nucleotide sequences published in the National Center for Biotechnology Information (NCBI) nucleotide database (www.ncbi.nlm.nih.gov/nuccore); and (b) titles and abstracts of publications indexed in the PubMed database (www.ncbi.nlm.nih.gov/pubmed). To date, EID2 has extracted information from >7 million sequences (and processed ≥ 100 million sequences) and >8 million titles and abstracts. EID2 imports the names of organisms and their taxonomic hierarchy from the NCBI taxonomy database (http://www.ncbi.nlm.nih.gov/Taxonomy/) and aligns them with an exhaustive collection of alternative names. In general, EID2 follows the NCBI definitions of “species” and “subspecies”, with unclassified and uncultured species being denoted as “no rank”.
The data of interest for the present study were associations of mammalian species (including humans) with different virus species, independent of location records. We considered a mammalian species to be host to a virus if at least one NCBI meta-dataset accompanying a published sequence detailed an association between the virus (or any of its subspecies or strains) and the host (or any of its subspecies), including detailed information about the sampling location (e.g., country/county where the association was recorded). We used this conservative approach rather than the full range of information collated from sequence records and text mining in order to reduce any possible bias from experimental infection studies. However, although we assume that sampling locations are most likely to be recorded as metadata for natural infection, we are aware that our dataset might include non-natural infections.
Virus species were assigned to genome type (DNA, RNA or other/unspecified) following NCBI taxonomy as used by EID2. Mammal species synonyms and taxonomic orders were standardized using the taxonomy of Wilson and Reeder (2005), the online version of IUCN Red List and Integrated Taxonomic Information System (ITIS; accessed May 2018). This revision enabled us to match the most recent host names to trait data.
Of the 724 non-human mammalian host species in our dataset, we considered 21 species as “domestic” (including the major commensal rodent species) and all others as “wildlife”. Domestic species were banteng (Bos javanicus), yak (Bos mutus), cow (Bos taurus), water buffalo (Bubalus bubalis), bactrian camel (Camelus bactrianus and Camelus ferus), dromedary (Camelus dromedarius), dog (Canis familiaris and Canis lupus), goat (Capra aegagrus), guinea pig (Cavia porcellus), wild ass (Equus africanus), donkey (Equus asinus), horse (Equus caballus), cat (Felis catus), guanaco (Lama guanicoe), house mouse (Mus musculus), rabbit (Oryctolagus cuniculus), sheep (Ovis aries), brown rat (Rattus norvegicus), black rat (Rattus rattus), pig (Sus scrofa) and vicugna (Vicugna vicugna). We constrained our domestic species selection to these major domestic species only to showcase possible differences in pathogen sharing, although we are aware that there are some additional species that might be considered to be domestic animals.
We generated four different measures of sampling effort for each mammalian host species, namely: (a) number of PubMed-indexed publications (summed over all associated virus species); (b) number of virus sequences recorded (summed over all associated virus species); (c) Shannon diversity of publication records, accounting for the proportional number of publications for each associated virus species; and (d) Shannon diversity of sequence records, accounting for the proportional numbers of sequence records for each associated virus species. For Shannon indices, larger values are linked to an overall larger number of records and a more even distribution of records among different virus species, that is, higher overall sampling coverage (Magurran, 2004). We generated these multiple indices as proxies of sampling intensity because the true sampling effort is not known. This is because records of species interactions in the literature are arguably “presence-only” records and rarely report the lack of interactions or the number of host individuals examined that would reduce the number of pseudo-absences in biotic interaction data (Little, 2004; Wells et al., 2013).
2.2 Mammalian host phylogeny and ecological trait data
A goal of this study was to assess whether variation in the phylogenetic and ecological similarities of mammalian species predicts patterns of virus sharing (i.e., pairwise phylogenetic and ecological distances that are calculated among all possible combinations of viable host species) and the proportion of zoonotic viruses (i.e., viruses infecting humans and at least one other animal species) associated with different host species. We gathered ecological trait data from the PanTHERIA (Jones et al., 2009) and EltonTraits v.1.0 (Wilman et al., 2014) databases to characterize all of the sampled mammals using a range of traits likely to impact on their suitability as hosts for viruses.
Selected traits were as follows: body mass, which is a key feature of mammals in terms of their metabolism and adaptation to environments; average longevity, litter size and the average number of litters per year, as demographic parameters that could be relevant for within-host dynamics of viruses; diet breadth (calculated as a Shannon diversity index based on the proportional use of 10 diet categories as presented in EltonTraits); range area, which we expect to affect the exposure to other mammalian host species; average temperature and average precipitation within the distribution of a host as an indicator of climatic niche; latitudinal centroid of distribution as an indicator of the general habitat and climate within which hosts are occurring across a gradient from tropical to polar environments; and habitat, as multiple binary indicators of whether a species uses forest, open vegetation and/or artificial/anthropogenic habitats. Information on specific habitat utilization was compiled from the International Union for the Conservation of Nature (IUCN) database (http://www.iucnredlist.org). Missing trait data were randomly imputed (as part of the Bayesian sampling approaches; see model codes in Supporting Information Appendix S1). We did not include a larger set of ecological traits in our analysis in order to avoid collinearity issues.
Phylogenetic relationships between sampled mammal species were estimated from a recent mammalian supertree (Fritz, Bininda-Emonds, & Purvis, 2009). We used this tree to compute pairwise phylogenetic distances based on a correlation matrix of phylogenetic branch lengths (Paradis, Claude, & Strimmer, 2004) and also a vector of phylogenetic distance to humans for all other mammalian host species. We also quantified pairwise ecological distance between sampled mammal species based on a generalized form of Gower's distance matrices (Gower, 1971) using weighted variables based on all of the ecological trait variables described above, following methods described by Pavoine, Vallet, Dufour, Gachet, and Daniel (2009). Phylogenetic and ecological distance matrices and the vectors of trait variables were scaled (dividing by the maximum for each distance matrix) such that all distance measures ranged from zero to one. Data formatting and analyses were conducted in R v.3.4.3 (R Development Core Team, 2017) and used the packages ape (Paradis et al., 2004) for phylogenetic distance calculations and ade4 (Dray & Dufour, 2007) for ecological distance calculations.
2.3 Statistical analysis
The primary focus of this paper was to explore which mammalian host species might be the most important for spreading viruses as a result of their sharing of viruses with others, and we were interested in the phylogenetic and functional diversity of host species infected by different virus species. We addressed these aims using three different statistical approaches, which we describe in detail in the Supporting Information (Appendix S1). In brief, we used the following approaches.
2.3.1 Centrality of host species in networks of virus sharing
We calculated eigenvector centrality (a generalization of degree, which is the number of connections a host species has to others in terms of virus sharing; eigenvector centrality accounts both for the degree of a host species and those of connected species, that is, it considers host species to be highly central if their connected species are connected to many other well-connected species (Bonacich & Lloyd, 2001)). Eigenvector centrality was strongly correlated with degree measures, betweenness centrality and closeness centrality (all Spearman’s r ≥ .76). Thus, we present only results from eigenvector centrality and acknowledge that because of collinearity, it is not possible to distinguish further between the different components.
We used the nonparametric Kruskal–Wallis test to assess whether the eigenvector centrality measures differed between wildlife and domestic species and among host orders. We applied Dunn's test for multiple comparisons (Benjamini & Yekutieli, 2001). To account for sampling variation that could bias centrality measures (larger sample sizes may increase the relative number of interactions reported for poorly sampled host species; Costenbader & Valente, 2003), we randomly removed subsets of interaction records from the adjacency matrix used for calculating centrality measures. For this, we varied the proportion of removed interactions between 5 and 30% in each of 200 iterations, following a uniform distribution. We used the relative proportion of publication and sequence numbers for each mammal–virus combination as two independent sets of probabilities of which interactions to remove. We then calculated centrality measured for each iteration and tested for consistency of results from subsets and the full dataset.
2.3.2 Hierarchical model of virus sharing among host species

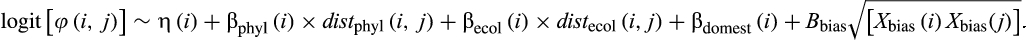
Here, η(i) is the species-specific intercept, which is modelled further with a hierarchical hyperprior as η(i) ~ N[Hη(order), ση(order)]; the hyperprior Hη accounts for the “average” virus-sharing probability of species from different orders, and the variance ση accounts for the deviation of species-level virus-sharing probabilities from the respective order-level hyperprior. The coefficients βphyl and βecol account for variation in virus sharing with increasing phylogenetic and ecological distance from i. The coefficient βdomest accounts for variation in virus sharing among all possible combinations between species classified as wildlife, domestic or human compared with pairs of wildlife–wildlife species (a five-level categorical variable). The coefficient vector Ɓbias accounts for variation in relationship to the four different proxies of sampling efforts described above, that is, it controls for sampling variation in the probabilistic model framework. Covariates from proxies of sampling efforts were generated as the square-rooted product of pairwise proxy variables. We fitted the model in a Bayesian framework with Markov chain Monte Carlo (MCMC) sampling in the software JAGS v.4.3.0, operated via the R package rjags (Plummer, 2016).
2.3.3 Hierarchical model of the proportion of zoonotic viruses carried by different host species
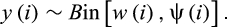
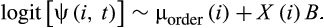
Here, µorder denotes the order-specific average according to the taxonomic order of species i, which was modelled with a Gaussian error structure and a common “average” hyperprior mean, that is, µorder ~ Ɲ(H, σ2). X is a matrix of the 17 species-level covariates (including phylogenetic distance to humans and the four proxies of sampling bias) described above, and B is a vector of corresponding coefficient estimates. This model accounts for sampling variation similar to the model of virus sharing (through variation partitioning among multiple covariates that are assumed to represent either the relevant biological processes or proxies of sampling bias). We fitted the model in a Bayesian framework in JAGS (Plummer, 2016).
3 RESULTS
Of 1,785 virus species associated with 725 different mammalian host species (including humans) in our dataset, 405 species (23%) have been recorded to infect humans. Of these, 138 species (34% virus species infecting humans) are recorded as zoonotic, and of the zoonotic species, 56 (41%) were recorded in wildlife but not in any domestic species, whereas 21 species (15%) were recorded in humans and domestic animals but not in any wildlife species; the remaining 61 zoonotic viruses were recorded in both wildlife and domestic species. In turn, 87 (5%) of all recorded virus species were shared by at least one domestic and one wildlife species without being associated with humans.
The virus species included 730 DNA virus species and 912 RNA virus species (73 classified as “others”), of which 24 (3% of DNA virus species) and 91 (10% of RNA virus species) were recorded as zoonotic. The overall network topography for DNA versus RNA viruses reveals distinct spread of these viruses among host species, mostly depicted by considerably lower virus sharing across orders of host species for DNA viruses (Figure 1).
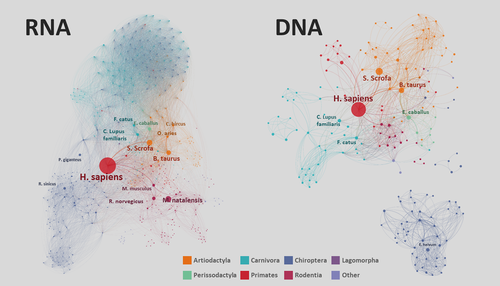
3.1 Centrality of host species in networks of virus sharing and spread
Eigenvector centrality measures were higher for domestic than for wildlife host species (Kruskal–Wallis χ2 ≥ 35, d.f. = 1, p < .01), indicating that domestic species were the most central species (after humans) in the entire mammal–virus association network based on current evidence. The 10 most central positions in the network of all virus species were occupied by Homo sapiens, Bos taurus, Sus scrofa, Ovis aries, Canis lupus, Capra hircus, Equus caballus, Felis catus, Bubalus bubalis and Mus musculus (in order of descending centrality).
Centrality measures also varied among the different taxonomic orders of host species (all Kruskal–Wallis χ2 ≥ 162.4, d.f. = 9, p < .01; Figure 2). Specifically, eigenvector centrality measures for all virus species were largest for wildlife species of the taxa Carnivora, Chiroptera, Artiodactyla and Primates compared with other taxa (Rodentia, Eulipotyphla and others) according to post-hoc multiple comparisons (Supporting Information Table S1). RNA viruses but not DNA viruses accounted for relatively larger centrality scores for Carnivora and Chiroptera (both Mann–Whitney U-test of group-level comparisons p < .01), whereas centrality scores calculated for RNA and DNA viruses appeared to be of indistinguishable ranks for Artiodactyla (Mann–Whitney U-test p = .52) (Supporting Information, Figure S1).
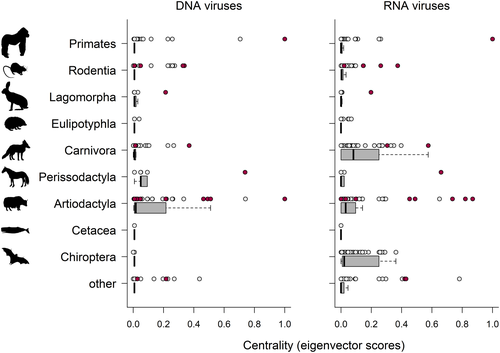
Centrality measures calculated from subsets of the underpinning adjacency matrix for all viruses, with 5–30% of interactions removed according to the number of published sequences and publications, revealed a 4-fold stronger decline in correlations for the number of published sequences than publications, but for all subsets, correlations with centrality measures from the full dataset remained reasonably high (i.e., all Spearman's r > .6 for centrality measures with ≤ 30% of interactions removed; Supporting Information Figure S2). For these data subsets, there were a total of 28 host species that emerged as the top 10 host species according to centrality measures calculated from data subsets (Supporting Information Figure S3). However, despite the uncertainty in which host species occupied the most central positions, the findings of significant larger centrality measures for domestic than for wildlife species held true for all subsets (all Kruskal–Wallis tests with χ2 ≥ 18.3, d.f. = 1, p < .01; Supporting Information Figure S2). Likewise, centrality measures varied among the different taxonomic orders for all subsets (all Kruskal–Wallis tests with χ2 ≥ 22.3, d.f. = 1, p < .01), with the same order showing the largest centrality measures as for the full dataset.
3.2 Virus sharing among host species
Analysis of virus-sharing patterns in a probabilistic hierarchical modelling framework confirmed the prominent role of domestic animals in virus sharing across the entire network. Wild mammalian host species were c. 5.7 times [95% credible intervals (CIs) of odds ratio 5–9.3] more likely to share virus species with humans and c. 4.2 times (odds ratio 4.9–5.5) more likely to share virus species with domestic animals than with any other wild species. Any pair of domestic species was c. 70 times (odds ratio 49.4–102.5) more likely to share viruses than any pair of two wildlife species. Humans shared DNA viruses c. 33 times (odds ratio 7–147) more often with any domestic species than DNA viruses were shared among any pair of two wildlife species, but we found no evidence that RNA viruses were shared more frequently by humans and any domestic species than among any pair of wildlife species (odds ratio 1–126).
We found the highest frequencies of sharing an RNA virus with any other mammalian species for species of the orders Chiroptera and Carnivora (averaging frequencies of .5–2% according to CIs of sharing RNA viruses with other species), whereas DNA virus-sharing frequencies were mostly < .2% (according to upper bounds of CIs except for the orders Perissodactyla and Cetacea, for which large CIs indicated imprecise estimates; Figure 3). For most host orders (except Cetacea) and both virus genome types, we found virus sharing to be more likely with closely related species (negative values for βphyl coefficients that depict increasing virus sharing for smaller phylogenetic distances among pairs of host species). Phylogenetic clustering of host species (which translates into higher phylogenetic host specificity for the viruses) was stronger for DNA viruses compared with RNA viruses shared by Primates, Carnivora, Artiodactyla and Chiroptera (Figure 3), signifying a general tendency for higher host specificity in terms of phylogenetic similarity for DNA viruses compared with RNA viruses. This tendency, however, was not true for viruses shared by Rodentia, because phylogenetic host specificity appeared to be relatively stronger for RNA than for DNA viruses associated with species from this order (Figure 3).

Notably, phylogenetic host specificity for RNA viruses shared by Primates was relatively low, suggesting more frequent host sharing with more phylogenetically distant host species than in other orders (Figure 3). We found species of the orders Primates, Carnivora, Artiodactyla and Chiroptera to share RNA viruses with any other hosts of larger functional distances than expected by chance, indicating low functional specificity of these viruses (positive values for βecol coefficients; Figure 3); however, functional distances among host species were generally less meaningful in describing patterns of virus sharing among pairs of host species than phylogenetic distances, as depicted by the smaller effect sizes (Figure 3). Virus sharing among host species increased with the four proxies of sampling bias for both DNA and RNA viruses (all CIs of odds ratios 1.03–3.03 except for the relationships of “Shannon diversity of publication records” with RNA virus sharing and “number of publications” with DNA virus sharing), indicating that sampling efforts impact the topography of currently known mammal–virus networks.
3.3 Proportion of zoonotic viruses in different host species
We found Primates to harbour the overall largest proportions of zoonotic viruses, with a group-level average of 51% (CI of 40%–63% for the respective µorder; Figure 4), followed by a slightly lower proportion of zoonotic viruses in Rodentia, Carnivora, Artiodactyla and Chiroptera (all respective µorder CIs ranging between 12 and 46%; Figure 4). The proportion of zoonotic viruses carried by domestic species was 1.8 times higher than in wildlife (odds ratio of 2.8 and CI of 1.8–4.3). RNA virus species accounted for the highest proportions of zoonotic viruses in all mammalian groups, averaging to 38% (CI of 15–64% according to hyperprior HRNA) compared with only 9% (CI of 2–24% according to hyperprior HDNA) of the DNA viruses in mammalian hosts being zoonotic.
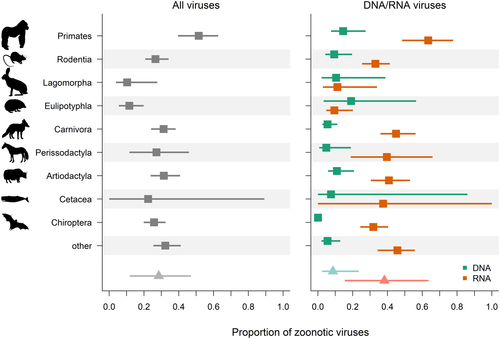
We found the proportion of zoonotic RNA viruses in different host species to increase with larger range area (odds ratio of 1.06–1.6). In contrast, there was no evidence that the proportion of zoonotic DNA viruses in different host species was linked to any species traits (all odds ratio estimates intersecting with one). The proportion of zoonotic RNA viruses was smaller for host species with higher Shannon diversity scores of sequence records (odds ratio of .6–.8), suggesting that more intensive sequencing efforts of a large range of these viruses has increased the discovery of viruses confined to non-human hosts.
The associations between host species from different mammalian orders and viruses from different families is illustrated in Supporting Information Figure S4, and data are presented in Supporting Information Table S2.
4 DISCUSSION
Pathogen spillover and the emergence of infectious diseases ultimately depend on how pathogens conquer eco-evolutionary barriers to infect novel hosts (Lloyd-Smith et al., 2009), but spatio-temporal variation in species interaction and pathogen transmission opportunities are proximately driven by host occurrences and community assembly (Canard et al., 2014; Stephens et al., 2016). It comes as little surprise, therefore, that globally pervasive mammal groups, such as bats and rodents, are often considered to share as many viruses with humans as do primates, our closest relatives (Calisher, Childs, Field, Holmes, & Schountz, 2006; Luis et al., 2013; Olival et al., 2017). Our study adds new insights into virus spread across mammalian communities. Specifically, we provide the strongest evidence to date that domestic animals are the most central species in mammalian host–virus interaction networks. We also find rather distinctive patterns of how DNA and RNA viruses are shared and spread among different mammalian groups, with bats and carnivores being most influential in spreading RNA viruses and playing only a minor role in spreading DNA viruses through the network. We emphasize the dominant role of domestic species in virus sharing, because domestication status strongly increases the chance of virus sharing among multiple mammalian hosts. Likewise, we also find domestic species to carry larger proportions of zoonotic viruses than wildlife species after accounting for phylogeny and other traits.
Our study concerns the contemporary pattern of virus sharing of mammal species rather than any specific co-evolutionary histories of host switching and origin of viruses. In many, perhaps most instances, this sharing indicates the possibility of cross-species transmission, either directly via contact or indirectly via air, soil, water, fomites or vectors. The exceptionally high virus sharing of humans and domestic animals with other mammalian species suggests that these species play a crucial role in spreading viruses, because frequent virus acquisition and dissemination is the most plausible explanation for such intensive virus sharing. This might reflect the wide geographical distribution and opportunities for contact with wildlife across biogeographical borders, given that domestic species are not particularly distinguished from wildlife in terms of ecological traits. In fact, opportunity for contact and community assembly have been shown in a number of studies to impact pathogen sharing and host shifting (Clark et al., 2018; Cooper, Griffin, Franz, Omotayo, & Nunn, 2012; Wells & Clark, 2019). Many pathogens, including viruses, can overcome species and environmental barriers to infect distantly related hosts and disperse across large geographical areas (Longdon, Brockhurst, Russell, Welch, & Jiggins, 2014; Wells, O'Hara, Morand, Lessard, & Ribas, 2015), although strong constraints in host shifting may also cause biogeographical structure in pathogen diversity and zoonotic disease risk (Murray et al., 2015; Poulin, 2010). Beside the large geographical ranges and diverse habitats encroached by domestic species, their large population sizes and high densities, which often exceed those of wildlife populations (Bar-On et al., 2018), could also contribute to host shifting and pathogen spread. This could be the case especially if large population sizes facilitate the opportunity for contact, virus amplification and diversification caused by more intensive within-population transmission or other factors, warranting future research.
Our findings of larger proportions of zoonotic RNA viruses compared with DNA viruses carried in different mammals is consistent with previous research (Cleaveland et al., 2001; Kreuder Johnson et al., 2015; Olival et al., 2017) and is in line with our finding that mammal species generally share RNA viruses with other hosts more frequently than they share DNA viruses. Here, we reveal, for the first time, that these two major groups of viruses are spread differently across entire networks of mammalian hosts, which is an important finding that remains largely unnoticed when looking solely at the species richness and propensity of zoonotic viruses carried in different wildlife species. Remarkably, Chiroptera and Carnivora hold central positions in terms of virus sharing with other species for RNA viruses only, whereas ungulates hold central positions for sharing both RNA and DNA viruses with other host species. In practice, these findings translate into a minor role of bats and carnivores for the spread of DNA viruses (and relatively low risk that DNA viruses will spillover from these species to humans). We also found that cattle (Bos taurus), pigs (Sus scrofa), horses (Equus caballus) and sheep (Ovis aries), which are globally the most abundant and economically important mammalian livestock species (Thornton, 2010), are among those species with the relatively highest centrality measures in terms of DNA virus sharing. Importantly, it should be noted that for all these species, the frequencies of sharing DNA viruses with other host species was considerably lower than for sharing RNA viruses regardless of centrality measures (as is also true for group-level estimates for different mammalian orders, as depicted in Figure 3). We thus emphasize that the aforementioned species have a relatively crucial role in spreading DNA viruses, whereas RNA viruses generally are much more frequently shared among mammalian host species. In this context, our model framework for analysing patterns in host sharing provides probabilistic estimates of the variation in the pairwise phylogenetic and functional similarities of infected versus uninfected host species as a signal of host specificity. This tool enables us to quantify host specificity of DNA versus RNA viruses in different groups of hosts, resulting in refined and community-wide measures of previously notified higher host specificity in DNA viruses compared with RNA viruses (Cleaveland et al., 2001; Geoghegan et al., 2017; Jackson & Charleston, 2004). Notably, the low functional host specificity of RNA viruses exhibited by viruses shared among hosts of Primates, Carnivora, Artiodactyla and Chiroptera (i.e., functional traits of pairs of host species infected by these viruses were larger than expected by chance) emphasizes their capacity to cross ecological species barriers during host-shifting events despite the overall tendency to infect phylogenetically related species.
The understanding of virological factors that ensure efficient virus replication and transmission within and among host species is in its infancy (Geoghegan, Senior, Giallonardo, & Holmes, 2016). Consequently, disentangling host or virus traits as drivers of the differential spread of DNA and RNA viruses among different mammalian orders is currently not possible and requires additional research. Possible working hypotheses as to why primates and ungulates are of relatively high central importance in sharing DNA viruses could be linked to mechanisms that enable efficient within-host virus replication and population-level transmission. At the same time, exploration of virus attributes of the major DNA virus families shared among these host species, namely Herpesviridae, Papillomaviridae and Adenoviridae (Supporting Information Figure S4), might help to explain why these viruses are more likely to be shared by primates and ungulates but are less likely to cross host species barriers with regard to bats and carnivores. Moreover, the strong links of some RNA viruses, such as the Bunyavirales, to arthropod vectors (Marklewitz, Zirkel, Kurth, Drosten, & Junglen, 2015) requires further research into the role of host–vector associations and other transmission modes for the spread of viruses.
We recognize several shortfalls in analysing database records of host–pathogen associations. First, any record of a virus species in a host relies entirely on targeted molecular screening. Certain research foci, such as the boost in coronavirus research linked to bats after the severe acute respiratory syndrome (SARS) pandemics (Drexler, Corman, & Drosten, 2014), may include a sampling bias difficult to capture when accounting only for publication or sequencing numbers as proxies for sampling bias, because the true presence/absence of viruses in non-target host species remains unknown. Undoubtedly, major research efforts are linked to viruses of public health relevance, whereas there is a dearth of systematic pathogen surveillance in wildlife (Tompkins, Carver, Jones, Krkošek, & Skerratt, 2015). Whether different sampling efforts for DNA and RNA are captured sufficiently by the proxies for sampling bias is unknown and warrants future research. Second, detection of a pathogen in any targeted host species depends on its prevalence in its host population and the number of sampled host individuals, but such information is not always available from collated database records. With sparse data, any direct interpretation of absolute numbers of species richness and interactions could instead reflect the observation process rather than true biological patterns and processes (Wells et al., 2013), and we are therefore not able to explore such important properties in our study. Network topologies can be also biased by sampling and data aggregation (Farine & Whitehead, 2015). We control for research effort in our analysis by accounting for variation in relationship to publications and sequencing numbers, as has been done previously (Gómez et al., 2013; Olival et al., 2017). However, as more complete data from systematic disease surveillance efforts becomes available, it will be desirable to improve such analysis to better distinguish true but undiscovered interactions from “false zeros” among other sources of bias. Compiling host–pathogen interactions from the literature and published evidence may also lead to “false positives”, such as interactions recorded from laboratory infection studies only; we minimized this error in our study by considering only interactions backed by molecular sequence records with information about sampling location in the metadata. The ongoing sophistication and broad-scale application of molecular screening methods for detecting pathogen species and identifying lineage variation might also lead to the discovery of unexpected and cryptic interactions among previously disconnected groups (Doña, Serrano, Mironov, Montesinos-Navarro, & Jovani, 2019). Finally, we are aware that amalgamating species-specific host–pathogen interactions into an N × N adjacency matrix, as used for some network statistics, comes at the cost of losing information about pathogen species identity, and thus the overall connectivity of host species can no longer be traced back to particular pathogen species. Overall, network connectivity and modularity are therefore community-level entities, whereas a focus on particular virus species would require more detailed analysis of underlying species-level interaction matrices.
Our work reveals the importance of domestication status and phylogenetic clustering on the importance of virus sharing among mammals, also showcasing the limited sharing of DNA viruses by bats and carnivores, in contrast to primate and ungulate species that readily share both RNA and DNA viruses. The emergence of new infectious diseases through pathogen spillover is a hierarchical process. Ecological factors that determine the opportunity for contact between different host species pave the way for cross-species transmission, host adaptation and subsequent within-host reproduction and transmission, which are then controlled largely by ecophysiological and genetic factors. Future work that better accounts for virus factors and host species community assembly might shed further light on why different types of viruses spread differently among phylogenetic and functional groups of mammals and foster better predictions of future disease emergence.
ACKNOWLEDGMENTS
Establishment of the EID2 database was funded by a U.K. Research Council Grant (NE/G002827/1) to M.B., as part of an European Research Area Networks Environmental Health award to M.B. and S.M.; subsequently, it has been developed further and maintained by Biotechnology and Biological Sciences Research Council (BBSRC) Tools and Resources Development Fund awards (BB/K003798/1; BB/N02320X/1) to M.B., and the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Emerging and Zoonotic Infections at the University of Liverpool in partnership with Public Health England and Liverpool School of Tropical Medicine. The views expressed are those of the authors and not necessarily those of the National Health Service, the NIHR, the Department of Health or Public Health England. S.M. is supported by the French ANR FutureHealthSEA (ANR-17-CE35-0003). M.W. acknowledges support from BBSRC and Medical Research Council for the National Productivity Investment Fund (NPIF) fellowship (MR/R024898/1).
Open Research
DATA ACCESSIBILITY
The data reported in this paper are deposited at Dryad (https://datadryad.org/stash/dataset/doi:10.5061/dryad.p2ngf1vmg).
REFERENCES
BIOSKETCH
As a team, the authors combine complementary interests in wildlife and disease ecology, including host–pathogen interactions, parasite biogeography, biodiversity, computational epidemiology, open and big data approaches and One Health. Their collective aim is to understand and predict the spread of pathogens from populations to communities in order to gauge and mitigate disease risk in times of global change.




