Decoupled erosion of amphibians’ phylogenetic and functional diversity due to extinction
[Correction added on 05 May 2020, after first online publication: the name of the 2nd author was misspelt and has been corrected in this version.]
Abstract
Aim
It has been argued that the loss of phylogenetic diversity (PD) from species extinctions will result in concomitant loss of functional diversity (FD). As a result, species extinction undermines not only unique evolutionary history, but also ecosystem function and stability. Using data from > 6,000 amphibian species globally, we assess the potential erosion of PD and FD as a result of extinction.
Location
Global.
Time period
From the present day to the next 100 years.
Major taxa studied
Amphibians (Anura, Caudata and Gymnophiona).
Methods
We simulated future biogeographical patterns of extinction based on International Union for Conservation of Nature (IUCN) threat status, and estimated the loss of PD and FD accordingly. We used null models to determine the extent to which extinctions may result in disproportional losses of PD and FD relative to random extinction. Using spatial regressions, we tested whether the loss of PD can predict the loss of FD.
Results
Although spatial patterns of current amphibian PD and FD were similar, extinctions did not retain this similarity. The magnitude of such a decoupling varied widely in space, with most of the global assemblages losing more FD than PD.
Main conclusion
This study challenges the assumption that extinctions should yield comparable loss of PD and FD. Species may not be equivalent in the amount of unique evolutionary history and ecological functions that would be lost if they become extinct. Designing conservation strategies based on a single dimension of biodiversity has the danger of leading to misinformed outcomes.
1 INTRODUCTION
Rapid loss of biological diversity from human actions has triggered a mass extinction during the Anthropocene (Barnosky et al., 2011; Ceballos et al., 2017). This might include the loss of species that play important ecological roles within communities (Boyer & Jetz, 2014; Fonseca & Ganade, 2001), owing to their unique traits (Faith, 2018) and evolutionary history (Veron, Davies, Cadotte, Clergeau & Pavoine, 2017). A common assumption is that closely related species share more similar ecologies than distant related ones (Wiens & Graham, 2005), thus protecting phylogenetic diversity (PD) may help to preserve functional diversity (FD) (Faith, 1992, 2008; Vane-Wright, Humphries & Williams, 1991; Veron et al., 2017; Winter, Devictor & Schweiger, 2013). The correlation between PD and FD is a topic of intense debate (Gerhold, Cahill, Winter, Bartish & Prinzing, 2015; Pavoine, Vela, Gachet, Bélair & Bonsall, 2011; Tucker, Davies, Cadotte & Pearse, 2018), with evidence both for it (Collen, Turvey, et al., 2011; Huang, Stephens & Gittleman, 2012; Redding, Dewolff & Mooers, 2010) and against it (Kelly, Grenyer & Scotland, 2014; Mazel et al., 2018; Oliveira et al., 2016). However, whether the loss of PD yields concomitant loss of FD remains poorly understood, therefore undermining our capacity to predict the impact of extinctions on evolutionary and ecological dimensions of biodiversity.
The magnitude of PD loss and FD loss from any species’ extinction ultimately depends on its shared evolutionary history and ecological similarity with extant species, respectively. Due to phylogenetic constraints on trait and niche evolution (Felsenstein, 1985; Wiens & Graham, 2005), the simplest scenario predicts that the loss of PD and FD should be coupled. For instance, the extinction of a species in an older monotypic clade with few relatives could have a greater impact on the loss of both PD and FD than the loss of a species in a recently diverged clade with many close relatives (Nee & May, 1997; Redding et al., 2010). This is because the former species might carry more distinct evolutionary history and ecological traits not found elsewhere than the latter species. However, different pathways exist that can lead to decoupled losses between PD and FD. For instance, a large portion of PD would be lost with the extinction of phylogenetically isolated species, but FD loss could be low if remaining species share similar ecological roles (i.e., functional redundancy, Rosenfeld, 2002). This can occur via convergent evolution (Winemiller, Fitzgerald, Bower & Pianka, 2015), or strong environmental selection (Li et al., 2018; Webb, Ackerly, McPeek & Donoghue, 2002), that expresses similar suites of traits among phylogenetically distant lineages. Conversely, high FD can be lost if some traits correlate strongly with extinction risk. Species with small geographical range size, low population density, long life span, slow reproductive rates and large body size are disproportionately more threatened (Collen, McRae, et al., 2011; Davidson, Hamilton, Boyer, Brown & Ceballos, 2009; Fritz, Bininda-Emonds & Purvis, 2009; Jenkins, Pimm & Joppa, 2013), and so the loss of these traits by extinction may disproportionately reduce FD relative to PD (Fritz & Purvis, 2010).
Studies concerning the loss of biodiversity by extinction often focus on single dimensions of biodiversity at a time (e.g., Batista, Gouveia, Silvano & Rangel, 2013; Boyer & Jetz, 2014; Jernvall & Wright, 1998; Jono & Pavoine, 2012), but there is clearly a need for more integrated approaches that account for multiple dimensions jointly (Brum et al., 2017; Cadotte, Davies & Peres-Neto, 2017; Cadotte & Tucker, 2018; Pollock, Thuiller & Jetz, 2017). Global analyses with mammals showed that the loss of PD, inferred by the extinction of threatened species in accordance to the International Union for Conservation of Nature (IUCN) Red List, did not match losses of FD estimated by variance in body mass (Davis, Faurby & Svenning, 2018; Fritz & Purvis, 2010). Such decoupling undermines the use of PD loss as a surrogate for FD loss. Although body mass is arguably an indicator of many facets of species’ ecology and life history, combinations of traits corresponding to multiple ecological dimensions should yield more holistic representation of FD (Kraft, Godoy & Levine, 2015; Petchey & Gaston, 2006). For instance, Tucker et al. (2018) found that the strength of the PD–FD relationship increases with increasing number of traits used in the FD measure.
PD theoretically reflects the aggregated expectation of multiple, including hard-to-measure, traits. Consequently, PD has been widely used as a proxy of FD. Furthermore, Faith (1992, 2018) argued that PD may also help to protect unknown FD of unanticipated future benefits for humanity, an outcome referred to as its “option-value”. In this study, we focus on the “proxy-value” of PD, not its “option-value”, both being important reasons for conserving PD (Faith, 2015; Forest, Crandall, Chase & Faith, 2015; Owen, Gumbs, Gray & Faith, 2019).
Here, we integrate phylogeny (Jetz & Pyron, 2018), traits (Oliveira, São-Pedro, Santos-Barrera, Penone & Costa, 2017), distribution and threat status (IUCN, 2018) for amphibians globally (6,097 species) to predict how extinctions will impact their PD and FD. We do this by first calculating current amphibian PD and FD, and then estimating the loss of PD and FD given probabilities that currently threatened species will go extinct in the near future (Mooers, Faith & Maddison, 2008). We assess the impact of extinctions on total amphibian diversity, as well as patterns of diversity loss across space according to species assemblage within grid cells. Understanding the consequences of extinctions on amphibian diversity is crucial for prioritizing conservation because they face high extinction pressure from human impacts, with 41% of species threatened with extinction (IUCN, 2018). Our study reveals that although patterns of current amphibian PD are FD associated, extinctions would result in strongly decoupled losses of amphibian PD and FD.
2 METHODS
2.1 Data
We used the most recent amphibian phylogeny (Jetz & Pyron, 2018) containing nearly all of extant amphibian diversity. This phylogeny represents an updated version of the Pyron and Wiens (2011) phylogeny, and incorporates publicly available molecular sequence data from earlier studies and from biodiversity collections. This phylogeny is available as a set of 10,000 supertrees, from which we obtained a single maximum clade credibility (MCC) tree for use in downstream analyses. The MCC tree was obtained using the maxCladeCred function from the R package phangorn v. 2.5 (Schliep, 2011). We compared the results obtained with this MCC phylogeny with those obtained with 100 phylogenies selected from the posterior distribution of 10,000 phylogenies from Jetz & Pyron (2018). That we used a MCC tree is unlikely to affect our results (Supporting Information Figure S1).
Trait data were taken from AmphiBIO (Oliveira et al., 2017). From this dataset, we selected the four most complete traits: body size (log-transformed), habitat (fossorial, terrestrial, aquatic and arboreal), number of clutches per year (log-transformed) and breeding strategy type (direct developer, oviparous, or viviparous/ovoviviparous). We imputed missing data using the nonparametric version of the random forest algorithm that considers allometric relationships among species traits (R package missForest v. 1.4, Stekhoven & Bühlmann, 2012). Given that amphibians exhibit a strong order-specific allometric relationship, we kept amphibian order (Anura, Caudata and Gymnophiona) as an imputation variable (Santini, Benítez-López, Ficetola & Huijbregts, 2018). We did not use phylogeny for imputing misinformation as it might artificially increase the correlation between phylogeny and traits. The random forest algorithm performs comparably to or better than its alternatives, even without phylogenetic information (Penone et al., 2014). We summarized traits using principal coordinates analysis (PCoA) with a Gower’s distance matrix (Gower, 1966) calculated on the complete dataset, so that total inertia and distance between the species was kept constant across analyses. We used the first three PCoA axes as traits, which accounted for 85% of the variation in the data. To assess the sensitivity of our analyses to imputation, we compared spatial patterns in FD obtained with the imputed dataset with those patterns obtained with a reduced dataset that included only species from which we had complete trait coverage (n = 3,486). We found strong correlation between FD values calculated with both datasets (Supporting Information Figure S2).
We obtained amphibian range maps from the IUCN (IUCN, 2018). We divided the world into a 2 × 2 equal area grid under a Lambert cylindrical equal area projection, and recorded species presence/absence in each grid cell. This resolution accurately represents species geographical ranges, richness and other macroecological patterns based on range map data, especially for less well known taxa such as amphibians (Hurlbert & Jetz, 2007). We define an assemblage as the group of species present in a grid cell. We discarded species absent from our phylogeny or trait data, as well species classified as extinct or extinct in the wild. Our final global dataset comprised 6,189 amphibian species distributed across 3,150 assemblages.
2.2 Simulating extinctions
A common approach when measuring biodiversity loss is to assume a scenario in which all threatened species go extinct. However, this depicts a very pessimistic and unrealistic scenario – it is unlikely that all threatened species will go extinct, nor will all non-threatened species survive. We follow Mooers et al. (2008) assignment of extinction probabilities to species based on their threat statuses from IUCN. We used 100-year extinction probabilities of .0001 for Least Concern (LC), .01 for Near Threatened (NT), .1 for Vulnerable (VU), .667 for Endangered (EN) and .999 for Critically Endangered (CR). We simulated extinctions by sampling each species 100 times based on its probability of remaining extant in a 100-year period (PropExtant = 1 – ProbExtinct).
2.3 Estimating the impact of extinctions on amphibian dimensions of diversity
Our analyses were carried out at global scale (across all amphibian species), and at regional scale (within each assemblage). The loss of amphibian dimensions of diversity was calculated for each of the 100 extinction simulations, and we report median and standard deviation (SD) values of diversity loss to facilitate interpretation.
We estimated PD using the Faith’s index (1992), which sums across all the branch lengths connecting species in a phylogeny, from tips to root. PD quantifies the amount of evolutionary history of a set of species in terms of millions of years. The extinction of a given species prunes the branch supporting that species, thus the amount of unique evolutionary history represented by that species. To estimate PD loss, we subtracted the PD of currently extant amphibians from the PD of survivor species (Faith, 2008).
A variety of FD indices has been proposed to summarize species distributions in trait space. Functional richness, perhaps the most commonly used FD metric, measures the volume of the trait space that is encapsulated by species (e.g., convex hull, Cornwell, Schwilk & Ackerly, 2006). A limitation of this metric is that it assumes trait spaces to be convex, thus being highly sensitive to outliers (e.g., extinction of species at the edge of the trait space) but invariant to extinctions in core areas of the trait space (Mouillot, Graham, Villéger, Mason & Bellwood, 2013). However, extinctions can occur in any position of the trait space, even creating holes, defined as unoccupied regions within this niche space (Blonder, 2016; Colwell & Futuyma, 1971), and we therefore do not use this index. Functional dispersion, another commonly used index, measures the overall clump of species in trait space (Laliberté & Legendre, 2010). A reduction of the number of species due to extinctions can increase trait dispersion, translating into more FD after extinction. This would be a counterintuitive pattern for the present study, and we therefore avoided the use of this index as well. We avoided the use of dendrogram-based FD indices since recent studies showed that these indices may lead to biased estimates and inaccurate ecological conclusions (Podani & Schmera, 2006; Poos, Walker & Jackson, 2009; Villéger et al., 2017).
To overcome these limitations, we used an algorithm that infers the shape and volume of high-dimensional objects via a thresholded kernel density estimate (hypervolume, Blonder, Lamanna, Violle & Enquist, 2014). Hypervolumes were constructed using the R package hypervolume v. 2.0.11 (Blonder, 2018), with a bandwidth estimated from the trait data (b = 0.02) using the method of Silverman (1992), and a quantile threshold of 95% for excluding outliers. Here, FD represents the volume of occupied trait space, assuming it may have a complex topography, with non-smoothed continuous shapes, rugged surfaces and filled with gaps (Blonder, 2016; Blonder et al., 2014). To estimate FD loss, we extracted the volume of the intersect area between the hypervolume of currently extant amphibians and the hypervolume of survivor species.
We used simulations to assess the usefulness of selected metrics for the purposes of this study. We simulated 1,000 phylogenies following a birth–death model, and traits were allowed to evolve in these phylogenies following a Brownian motion model (Supporting Information Appendix S1). The number of species in simulations followed a distribution akin to the observed species richness across global amphibian assemblages (ranging from 5 to 100 species). We found strong correlation between PD and hypervolumes obtained from simulations (Pearson’s r = .93), thus a decoupling between these metrics is unlikely to result from a spurious artifact of the different mathematical proprieties of each metric. Importantly, this correlation was much stronger than between PD and functional richness (Pearson’s r = .35) or functional dispersion (Pearson’s r = .14). We further tested whether FD metrics can identify extinctions in the centre of trait space. As expected, hypervolumes identified holes in trait space, whereas convex hulls and functional dispersions did not, confirming the utility of hypervolumes for this study. Detailed results and reproducible code used for running simulations are available in Supporting Information Appendix S1.
2.4 Accounting for data deficiency
Approximately a quarter of all amphibians lack sufficient information to be assessed based on IUCN criteria, and are thus classified as Data Deficient (DD) (IUCN, 2018). We use the k-nearest neighbour imputation algorithm (kNN) to predict DD threat status in order to include them in our analyses (Bland, Collen, Orme & Bielby, 2015; Howard & Bickford, 2014). The kNN method based on species ranges was shown to perform better than several other methods, including random forest (Veron et al., 2016). Therefore, we decided not to use random forests for imputing threat status as we did for imputing traits. Species geographical range sizes were derived from IUCN range maps. kNN imputations were performed using the R package VIM v. 4.8 (Templ, Alfons, Kowarik & Prantner, 2017), using the six nearest neighbours for imputations (Veron et al., 2016). Details on imputation accuracy are provided in Supporting Information Appendix S2. The predicted threat statuses used for DD amphibians are available in Supporting Information Appendix S3.
2.5 Null models
We used randomizations to determine whether the loss of amphibian PD and FD would be greater or smaller than expected under random extinctions. This was done by shuffling the threat status among species 100 times keeping total number of predicted extinctions constant, and recalculating each metric at each run. For global scale analyses, the shuffling of threat status was done among all species in the global pool, whereas for assemblage scale analyses we shuffled the threat status of species within each assemblage. Observed PD loss and FD loss were compared with their respective null expectation using the formula of standardized effect size (SES) [SES = observed – mean (null)/SD (null)]. Positive SES indicates higher loss of PD or FD than the average expected by random, while negative SES indicates lower loss of PD and FD than the average expected by random.
Metrics of PD and FD are often closely associated with species richness, and interpretation of these metrics would benefit from statistically controlling for this association. We compared extant PD and FD values with random values expected given species richness. Here, we randomly sampled the observed number of species in each assemblage 100 times, and recalculated PD and FD values during each interaction. To avoid comparisons of historically different assemblages, we used biogeographical realms to inform the species pool (Holt et al., 2013). We contrasted observed PD and FD values against their respective null distribution using SES. A positive SES indicates a higher PD or FD than expected by random, while a negative SES indicates a lower PD or FD than expected by random.
2.6 PD loss as proxy of FD loss
We modelled FD loss as a function of PD loss to test the validity of one dimension of diversity being a surrogate for the other. First, we fitted non-spatial regressions using ordinary least-square (OLS) models. OLS models contained significant spatial autocorrelation in their residuals, as investigated using Moran’s I (Legendre, 1993). In order to control for spatial autocorrelation present in the data, we fitted simultaneous autoregressive (SAR) models, which assume that the response variable is a function of both the explanatory variable and neighbouring locations (Kissling & Carl, 2008). As recommended by Kissling & Carl (2008), we specified neighbourhood distances using a row-standardized coding scheme for the spatial weights matrix. SAR models significantly removed spatial autocorrelation (Supporting Information Figure S3). To assess the explanatory power of the SAR models, we calculated pseudo-R2 values using Nagelkerke’s formula (Nagelkerke, 1991). Pseudo-R2 describes the variance explained by both predictor variables and spatial structure, which we partitioned to extract the pure effect of predictors (PD or PD loss) (Legendre & Legendre, 1998). We scaled all variables to a mean of 0 and a variance of 1 prior to analyses to make model coefficients comparable. Spatial analyses were implemented using the software sam v. 4 (Rangel, Diniz-Filho & Bini, 2010).
3 RESULTS
3.1 Global scale diversity loss
Our models indicate that over 32% (SD = ±0.33) of all amphibians (median = 1,979; SD = ±20) would be lost within the next 100 years. The extinction of these species eroded 26,046 Myr (SD = ±420) of unique evolutionary history, which corresponds to 20.4% (SD = ±0.33) of all amphibian PD and 5.3% (SD = ±0.55) of their FD (4PD:1FD ratio) (Figure 1). The imminent disappearance of at risk amphibians would remove far more of each dimension of diversity than expected by random extinctions (Figure 1), leaving a large PD and FD debt as unique lineages and trait diversity are at risk of becoming completely lost in the near future.
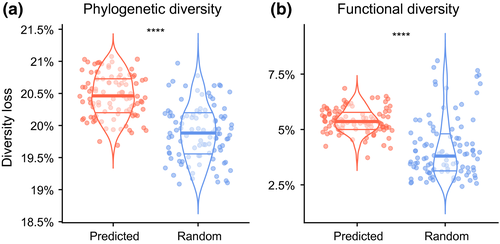
3.2 Regional scale diversity loss
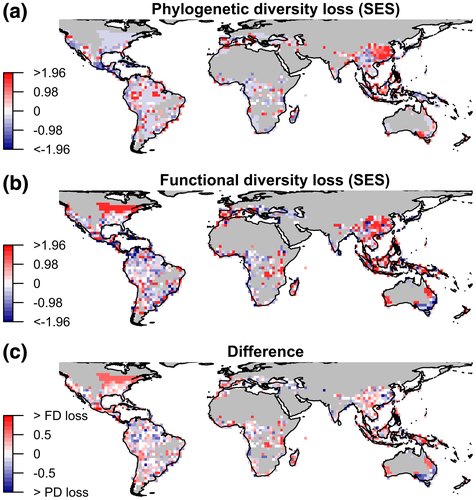
The loss of amphibian dimensions of diversity varied widely in space (Figure 2), regardless of the richness of extant species in assemblages (Supporting Information Figure S4 and Table S1). The loss of species richness was closely associated with PD loss within assemblages [standardized coefficient (std. coeff.) = .91, R2 = .85, p-value < .001], but not with FD loss (std. coeff. = .22, R2 = .4, p-value < .001) (Figure 2). Moreover, hotspots of species richness loss were mostly congruent with those of PD loss, but not with those of FD loss (Supporting Information Figure S5). Assemblages would lose on average more of their FD (7.8%) than PD (3.6%) (2.2FD:1PD), a pattern opposite to what was observed in the global scale assessment. Regions at risk of losing high proportions of both PD and FD were concentrated in Central America, the Caribbean islands, the Andes and northern Amazon of South America, eastern China, Southeast Asia and eastern Australia (Figure 2).

Our results revealed large geographical variation concerning the departure of PD loss and FD loss from random extinctions (Figure 3). The loss of PD and FD did not differ from random in most assemblages (denoted by a distribution of SES values centred on zero; Supporting Information Figure S6), but when they did they were more positive than negative, indicating higher loss of PD and FD than random (Figure 3; Supporting Information Figure S7). While all major regions of the world contained a mix of higher and lower loss for PD and FD, in most assemblages FD losses exceeded PD losses (Figure 3c), indicating elevated trait uniqueness in imperilled species. Exceptions were located mostly in the Neotropics and eastern Australia, where assemblages lost more PD than FD (Figure 3c).
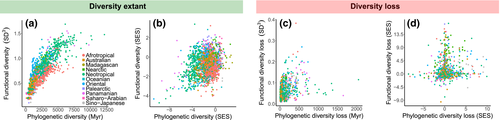
The PD and FD of currently extant amphibians were positively associated and followed an asymptotic relationship [Figure 4a; second-order polynomial function: R2 = .9, Akaike information criterion (AIC) = 524, p < .001; linear function: R2 = .87, AIC = 735, p < .001]. This positive association remained even after controlling for richness (Figure 4b). However, projected extinctions broadly decoupled the loss of PD and FD (Figure 4c,d), indicating that the loss of evolutionary history responded independently from the loss of ecological functions. Spatial models confirmed that the predictive power of PD on FD was greater than the predictive power of PD loss on FD loss (Table 1). These results again held after controlling for richness (Table 1, Figure 4d).
| Model | Std. coeff. | Pseudo-R2 | |
|---|---|---|---|
| Predictor + space | Predictor only | ||
| Extant diversity | |||
| FD ~ PD | .7 | .79 | .65 |
| FD SES ~ PD SES | .35 | .23 | .1 |
| Diversity loss | |||
| FD loss ~ PD loss | .34 | .23 | .12 |
| FD loss SES ~ PD loss SES | .11 | .27 | .02 |
Note
- Std. coeff. = standardized coefficient. The predictive power of models for extant diversity are much higher than the ones for diversity loss. Spatial models were calculated using raw PD and FD values, as well as using standardized effect sizes (SES) of these same metrics. Spatial models were fitted with simultaneous autoregressive models with the error term. Pseudo-R2 was partitioned into the variance explained by predictor variable and the combination of predictor variable + space. All std. coeffs. are significant (p-values < .05).
4 DISCUSSION
Understanding how extinctions may erode evolutionary and ecological dimensions of diversity is fundamental for conservation planning, especially when one dimension is used as a surrogate for another (Cadotte & Tucker, 2018; Devictor et al., 2010; Hidasi-Neto, Loyola & Cianciaruso, 2015; Mazel, Mooers, Riva & Pennell, 2017; Sobral et al., 2014). If shared evolutionary history among species infers ecological similarity (Felsenstein, 1985), then losing PD might result in the loss of FD (Faith, 2008; Forest et al., 2015; Nee & May, 1997; Veron et al., 2017). Our results indicate a weak association between the loss of PD and FD (Figure 4), therefore challenging the notion that PD loss is an appropriate surrogate of FD loss. Interestingly, this decoupling occurred even though a strong correlation exists between extant amphibian PD and FD across assemblages globally (Figure 4).
Although highly threatened (Brum et al., 2013; Pounds, 2001; Sodhi et al., 2008), analyses of projected impacts by extinctions on amphibian diversity are scarce. Batista et al. (2013) simulated the extinction of threatened New World anurans and found a spatial pattern in PD loss that closely resembles our results. However, as a consequence of knowledge gaps regarding traits, studies on amphibian FD have had narrow taxonomic and geographical scope (Ernst, Linsenmair & Rödel, 2006; Ochoa-Ochoa et al., 2019; Tsianou & Kallimanis, 2016). We provide the first global estimate of current patterns, and predicted future loss, of amphibian PD and FD. We found that current amphibian PD and FD are related to each other globally (Figure 4a,b), consistent with the hypothesis that closely related species are ecologically similar (Faith, 1992; Wiens & Graham, 2005). This association remains after controlling for richness using SES. Further support for this association comes from strong phylogenetic signal in the traits used for calculating FD (Supporting Information Table S2). However, there is little evidence that losses in PD and FD by extinctions will retain this similarity (Figure 4c,d). The lack of congruence between the loss of amphibian PD and FD indicates that species vulnerable to extinction vary in the magnitude of shared ancestry and functional redundancy with survivors. Importantly, this mismatch is geographically uneven, with some regions losing more FD than PD, whereas the opposite is expected for other regions.
We found that, for most worldwide amphibian assemblages, extinctions are likely to impact more of their current FD than PD (Figure 3c). This suggests threatened species carry more unique traits, or hang in shorter phylogenetic branches, relative to coexisting species. Previous studies of amphibians have shown that some traits correlate strongly with high threat risk [e.g., large body size, aquatic and arboreal habitat, viviparous reproduction, and small range size (Foden et al., 2013; Sodhi et al., 2008)]. A higher than expected global FD loss (Figure 1) indicates that threatened amphibians occupy a unique trait space. The loss of these species would greatly reduce variance among traits, leaving a high FD loss. Because amphibians show generally low dispersal ability and small range sizes, more diverse assemblages may have many closely related species, inducing phylogenetic redundancy and potentially contributing to a lower loss of PD than FD. Indeed, our data indicate assemblages with high richness of small-range species (range size < 200 km2) lost more species (Supporting Information Figure S8; Pearson’s r = .45, p-value < .001), but less PD than expected by chance (Supporting Information Figure S9; Pearson’s r = −.21, p-value < .001).
However, the Neotropical and eastern Australia regions were an exception for this pattern. Assemblages in these regions would lose more of their PD than FD, suggesting functional redundancy among species (Figure 3c). The Neotropics harbour the highest amphibian diversity and endemism globally, and both the Neotropical and eastern Australia regions concentrate a high number of threatened species (Jenkins et al., 2013). Recent phylogenetic analyses revealed that species-rich amphibian assemblages are composed of many old families that diversified quickly (Jetz & Pyron, 2018; Pyron & Wiens, 2013). The Neotropics hold highly evolutionary distinct species (Supporting Information Figure S10), thus their extinction would contribute to a high PD loss. Yet, rapidly diversifying amphibian clades share a greater number of threatened species compared to slowly diversifying clades (Greenberg & Mooers, 2017). If rates of trait evolution lag behind rates of lineage diversification, as in non-adaptive radiations (Kozak, Weisrock & Larson, 2006), it is possible that extinctions will overwhelmingly erode PD while maintaining FD due to high functional redundancy. This hypothesis is consistent with the observed asymptotic relationship between amphibian PD and FD, which is mainly driven by Neotropical assemblages (Figure 4a).
The relative impact of extinctions upon amphibian PD and FD varied with the scale of our analyses. At the global scale (across all 6,189 species considered in this study) amphibians lost more PD than FD (4PD:1FD ratio), whereas at the assemblage scale [2 ×2 grid cells, considered an appropriate resolution for the data (Hurlbert & Jetz, 2007)] they lost on average more FD than PD (2.2FD:1PD ratio). It is important to note, however, that any loss of PD or FD to extinction is relative to the survivors in the species pool. At the global pool, species from different assemblages overlap in trait space, resulting in higher functional redundancy relative to that at the assemblage scale. This high redundancy ultimately increases the chance that an extinct species will be replaced by a functionally equivalent one, with the loss therefore being less impactful to FD. On the other hand, in the assemblage pool, because amphibians in general have low vagility, diversifying lineages could remain within the same assemblage over time. This would result in high phylogenetic redundancy and potentially translate into less PD loss relative to FD loss. These seemingly contrasting results complement, rather than contradict, each other, allowing a more complete inference of the processes that predominate across different scales.
The poor support for the “proxy-value” of PD loss should not undermine the conservation importance of PD. Conserving PD may help to protect the diversity of evolutionary histories, but also multiple hard-to-measure traits that may be beneficial to society – referred to as the “option-value” (Amado, Bidau & Olalla-Tárraga, 2019; Faith, 2008, 2018; Forest et al., 2015; Owen et al., 2019). Any estimate of FD reflects the choice of traits, which is often arbitrary and determined primarily by data availability. We used traits that might link to ecosystem functions and services amphibians perform in ecosystems, and have been used in other studies with amphibians (Ochoa-Ochoa et al., 2019; Oliveira & Scheffers, 2019; Rapacciuolo et al., 2019). For instance, burrowing behaviour alters soil properties, body size is associated with feeding habits, and reproduction strategy may reflect energy and matter flux (Wells, 2007). Additional levels of detail in amphibian traits may change our representation of FD (Tucker et al., 2018) but high-resolution data remain currently unavailable.
In facing the current biodiversity crisis, conservation biologists have growing concerns over the degree to which future extinctions would erode evolutionary and ecological dimensions of diversity (Cadotte & Tucker, 2018; Davies, 2015; Mazel et al., 2016; Vane-Wright et al., 1991; Veron et al., 2017). Because our simulated extinction scenarios eroded PD and FD independently, we conclude that conserving one dimension of biodiversity as surrogate for another should be avoided. The loss of PD prunes the Tree of Life, and hinders the potential for lineages to evolve and adapt (Forest et al., 2015; Vane-Wright et al., 1991), whereas the loss of FD drives functional homogenization, which potentially reduces the goods and services from ecosystems that benefit humans (Fonseca & Ganade, 2001; Oliver et al., 2015; Petchey & Gaston, 2002). Prioritizing PD is generally a reasonably good method for conserving FD (Mazel et al., 2018), but the loss of one dimension may not yield concomitant loss of the other (Davis et al., 2018; Fritz & Purvis, 2010). Conservation attention should be directed towards regions with high likelihood of losing large portions of their PD and FD. However, a decoupling in the loss of PD and FD will require case-specific conservation strategies in different regions to secure the protection of the biodiversity dimension of interest (Cadotte & Tucker, 2018).
DATA ACCESSIBILITY
Distribution data are available through IUCN (https://www.iucnredlist.org/). Phylogenetic data are available through the Vert Life project (https://vertlife.org/). Trait data are available through the AmphiBIO database (https://figshare.com/articles/Oliveira_et_al_AmphiBIO_v1/4644424). Predicted threat status for Data Deficient species is available at the Supporting Information Appendix S3.
ACKNOWLEDGMENTS
This work was funded by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CAPES) and the Science without Borders grant Programa Professor Visitante do Exterior 018/2012 and the National Science Foundation Dimensions grants #DEB-1136586 and 1136588. BFO thanks scholarships provided by CAPES and Science without Borders. GCC thanks the Conselho Nacional de Desenvolvimento Científico e Tecnológico grants #302776/2012-5 and 201413/2014-0. BRS acknowledges financial support provided by the University of Florida and the Institue of Food and Agricultural Sciences as well as the National Institute of Food and Agriculture.
AUTHOR CONTRIBUTION
BFO and GCC planned the research. BFO performed the analyses. BFO wrote the first draft. All authors contributed substantially to the writing.
REFERENCES
BIOSKETCHES
Brunno F. Oliveira is interested in macroecology and community ecology. His research integrates phylogenies, maps and traits to understand the mechanisms that generate biodiversity gradients, and how humans disrupt these gradients.
Brett R. Scheffers is interested in biological organization and processes in space and time. Brett’s interests include multidimensional species distributions, community assembly/disassembly, ecophysiology, ecological scaling rules, and thermal complexity of landscapes. He uses these concepts to assess species and habitat vulnerability and resilience under novel climates and human disturbances.
Gabriel C. Costa is an Assistant Professor at Auburn University at Montgomery. His research focuses on understanding what determines the spatial distribution of biodiversity. He is interested in disentangling the roles of ecological and evolutionary drivers of biodiversity in different taxonomic groups and from local to global scales. He is also motivated by using his findings to create instrumental knowledge that can then support conservation decisions.




