Diversity regulation at macro-scales: species richness on oceanic archipelagos
Abstract
Aim
Understanding the mechanisms that generate diversity patterns requires analyses at spatial and temporal scales that are appropriate to the dispersal capacities and ecological requirements of organisms. Oceanic archipelagos provide a range of island sizes and configurations which should predictably influence colonization, diversification and extinction. To explore the influence of these factors on archipelagic diversity, we relate the numbers of native and endemic species of vascular plants, birds, land snails and spiders – taxa having different dispersal capabilities and population densities – to the number and sizes of islands in the major oceanic archipelagos of the globe.
Location
Fourteen major oceanic archipelagos of the globe.
Methods
Species richness was collated for native and endemic species in each archipelago. We used linear mixed effect models to quantify the influence on diversity of total area, number of islands, isolation and latitude. We then applied process-based modelling in a Bayesian framework to evaluate how speciation, colonization and extinction are influenced by characteristics of archipelagos associated with species richness, i.e. area, isolation and number of islands.
Results
We found parallel scaling of species richness among taxa with respect to total area and number of islands across groups. The process-based model supported effects of isolation on colonization and of area and number of islands on extinction rates, with the scaling exponents mostly similar across taxa. Data are consistent with a range of scaling exponents for speciation rate, implying that those relationships are difficult to infer from the data used.
Conclusions
We demonstrate an unexpected parallel scaling of species richness of four taxa with area and number of islands for the major oceanic archipelagos of the globe. We infer that this scaling arises through similar effects of the physical characteristics of archipelagos on extinction, colonization and speciation rates across these disparate taxa, indicating that similar mechanisms have created variation in diversity.
Introduction
Although theories in biogeography and macroecology have often emerged from observations of repeated patterns in nature (MacArthur, 1972), patterns themselves rarely provide substantial insight into underlying processes (Rahbek, 2005; Ricklefs, 2007). Indeed, a general consensus about the causes of global patterns of geographic variation in the number of species has yet to develop (Ricklefs, 2004).
The interpretation of large-scale species diversity patterns is dominated by a fundamental conceptual dichotomy, which contrasts (1) environmental limits to regional diversification and the number of locally coexisting species with (2) the influence of regional geography and history independent of local environmental conditions (Hutchinson, 1959; MacArthur, 1972; Ricklefs, 1987, 2007; Rosenzweig, 1995; Rabosky, 2009; Rabosky & Glor, 2010; Cornell, 2013). As multiscale palaeontological, biogeographic, genetic and ecological data have become more readily available, it is increasingly evident that these alternatives are not mutually exclusive. Large-scale patterns of diversity represent a balance between ecological and historical processes, and, although influenced by ecological conditions, the regional relationship between diversity and ecological space is not constrained to a particular environmentally determined relationship (see Brown et al., 2001; Ricklefs, 2004; Rahbek, 2005; Phillimore & Price, 2008; Rabosky & Glor, 2010; Kisel et al., 2011; Cornell, 2013; Rabosky, 2013; Economo et al., 2015). A key challenge to understanding the mechanisms that create diversity patterns is to frame analyses at the appropriate spatial and temporal scales.
Biotic provinces are self-contained areas within which diversity primarily reflects a balance between speciation and extinction, with colonization from the outside having a minor role. Accordingly, biotic provinces are appropriate spatial and evolutionary units for interpreting the causes of large-scale diversity patterns (Rosenzweig, 1995). Oceanic archipelagos are particularly useful biotic provinces for the analysis of diversity (Triantis et al., 2008; Cabral et al., 2014; Patiño et al., 2014; Valente et al., 2014). Being of volcanic origin and formed over oceanic plates, they have never been connected to continental landmasses and are among the most isolated biological systems of the globe. Moreover, most oceanic islands present a typical developmental sequence from youth, to maturity, old age and eventual disappearance under the ocean surface, giving oceanic archipelagos a similar temporal dynamic (Whittaker & Fernández-Palacios, 2007; Whittaker et al., 2008; Cameron et al., 2013) (Fig. S1 in Appendix S1 in Supporting Information). Finally, these archipelagos exhibit high endemism for most taxa, even the most mobile organisms, arising from a typically small number of colonizing species. For example, of the 29 species of land birds of the Galápagos, 83% are endemic to the islands; all seven bird species of Tristan da Cunha are archipelagic endemics. For less mobile taxa, such as land snails, endemism is high across all the major oceanic archipelagos, reaching up to 99% in Hawaii (e.g. Gillespie & Clague, 2009).
Here we used a two-stage approach to investigate the diversity of indigenous, native (indigenous but not endemic) and endemic species of vascular plants, birds, land snails and spiders of the 14 major volcanic archipelagos of the globe (Table 1 & Table S1 in Appendix S2). These four taxa differ greatly in their ecological requirements, dispersal abilities and typical population sizes, yet they occur on all oceanic archipelagos and are therefore good candidates for identifying commonalities and differences across spatial scales of biological organization (Whittaker & Fernández-Palacios, 2007; Gillespie & Clague, 2009).
| Archipelago | Indigenous | Archipelagic endemics | |||||
|---|---|---|---|---|---|---|---|
| Plants | Birds | Snails | Plants | Birds | Snails | Spiders* | |
| Azores | 230 | 23 | 104 | 86 | 1 | 74 | 24 |
| Canary | 1254 | 78 | 260 | 585 | 10 | 232 | 300 |
| Comoros | 937 | 47 | 146 | 500 | 16 | 100 | 45 |
| Galápagos | 549 | 29 | 100 | 236 | 24 | 94 | 88 |
| Gulf of Guinea | 903 | 49 | 110 | 107 | 34 | 83 | 92 |
| Hawaii | 1166 | 102 | 757 | 1024 | 98 | 752 | 162 |
| Juan Fernández | 207 | 12 | 40 | 121 | 6 | 20 | 28 |
| Madeira | 754 | 30 | 188 | 150 | 4 | 168 | 58 |
| Marquesas | 331 | 22 | 99 | 161 | 11 | 75 | 27 |
| Mascarene | 1224 | 60 | 200 | 749 | 51 | 180 | – |
| Northern Marianas | 180 | 5 | 30 | 21 | 0 | 12 | 3 |
| Samoa | 765 | 32 | 94 | 252 | 9 | 60 | 47 |
| Society | 575 | 26 | 161 | 250 | 16 | 136 | – |
| Tristan da Cunha | 91 | 7 | 15 | 45 | 7 | 15 | 4 |
- *For the archipelagic endemic spider species, data were available for 12 out of the 14 archipelagos considered.
First, we used linear mixed effect models in a regression framework to quantify the influence on diversity of potentially important macroecological drivers: total area, number of islands, isolation, geological age and latitude. At the archipelagic scale, how total area is partitioned among individual islands and how the islands are arranged spatially is likely to affect speciation rates, extinction rates and, ultimately, species richness (e.g. Kisel et al., 2011; Cabral et al., 2014). Second, to disentangle ecological and evolutionary mechanisms underlying patterns identified during the first descriptive step, we developed a process-based modelling framework in which species richness is a steady-state outcome of underlying processes: anagenetic and cladogenetic speciation, extinction and colonization. Our understanding of biodiversity has been advanced recently by the description, parameterization and empirical evaluation of biodiversity dynamics models that are built from simple assumptions (e.g. Gravel et al., 2011; Rosindell & Phillimore, 2011; Valente et al., 2014). In our framework, features of archipelagos, such as area and number of islands, affect the steady-state endemic and native richness through their influence on underlying processes. We took a Bayesian approach to inferring whether characteristics of archipelagos, as well as those of different taxa, influence richness patterns through their effects on underlying rates of colonization, extinction and speciation. We demonstrate an unexpected parallel scaling of species richness with area for birds, spiders, land snails and plants, regardless of variation in their ecology and propensity for colonization and dispersal within archipelagos. Our results suggest that this parallel scaling arises as a result of the similar effects of physical archipelagic characteristics on biogeographic rates across disparate taxa.
Methods
Archipelagos and physical attributes
We examined 14 oceanic archipelagos distributed across the globe (Table 1 & Table S1 in Appendix S2). The four taxa considered – land birds, vascular plants, spiders and land snails – are present on all the main oceanic archipelagos but have distinct ecological requirements and features. For each archipelago we recorded (1) latitude, (2) isolation (distance to the closest source area for colonists; see Rosindell & Phillimore, 2011), (3) maximum elevation, (4) maximum geological age for each of the archipelagos, defined as the age of the oldest island among the currently existing islands, (5) land area of the archipelago, and (6) number of islands constituting the archipelago. The configuration of islands varies greatly through time; sea level minima during the Pleistocene produced connections between some adjacent islands, turning them to single islands, while volcanism can both join and sometimes subdivide island areas (Whittaker & Fernández-Palacios, 2007; Carracedo & Tilling, 2003; Grant & Grant, 2008; Figs. S1 & S2 in Appendix S1 and Appendix S2; for further details see Cameron et al., 2013). As a simple description of archipelago dynamics, we considered previously connected islands in each archipelago as single islands (see below and Table S1 in Appendix S2).
The main sources of data were the UNEP Islands Directory (http://islands.unep.ch/) and several data compilations (Steadman, 2006; Whittaker & Fernández-Palacios, 2007; Whittaker et al., 2008; Gillespie & Clague, 2009; Rosindell & Phillimore, 2011; Cameron et al., 2013; Weigelt et al., 2013; Cabral et al., 2014; see also Appendix S4). For most of the archipelagos, if not all, the maximum geological age exceeds the age of the current islands. For example, the maximum geological age for the Canary Islands is estimated as 68 Ma, whereas the oldest current island in the archipelago, Fuerteventura, is only 20 Ma (Fig. S1A in Appendix S1). Although some lineages present on the archipelagos have histories that extend beyond the maximum age of the current islands (e.g. Givnish et al., 2009), the maximum age of the existing islands of an archipelago can be considered as a conservative reference point for the time over which biological processes have established modern diversity patterns (Price & Clague, 2002; Whittaker et al., 2008). All the archipelagos have maximum geological ages exceeding c. 3 Ma.
Species richness
For land birds, we included only species described as breeding, or probably breeding, on a given archipelago; for plants and land snails, we included all species considered indigenous or probably indigenous. Because species assemblages of these taxa on most oceanic islands have been affected by historic and pre-historic human activities (Olson & James, 1984; Steadman, 2006; Whittaker & Fernández-Palacios, 2007; Gillespie & Clague, 2009), we included both extant species and historically or prehistorically extinct species (Steadman, 2006) whenever such information was available. Olson & James (1984) noted that the pre-human bird faunas of the three main Atlantic archipelagos, i.e. the Azores, Madeira and Canary, are generally unknown (but see Sánchez-Marco, 2010), and so we also ran the analyses without them. However, excluding these faunas provided similar results (see Results and Appendix S3). Our sources of data on species richness were several published compilations (Steadman, 2006; Whittaker & Fernández-Palacios, 2007; Whittaker et al., 2008; Gillespie & Clague, 2009; Givnish et al., 2009; Sánchez-Marco, 2010; Rosindell & Phillimore, 2011; Cameron et al., 2013; Cabral et al., 2014; see also Appendix S4) and personal contacts with local experts (see Acknowledgements). For spiders, data were available for 12 archipelagos out of the 14 considered for the other three taxa, and only for the archipelagic endemics. Although different sampling and taxonomic efforts were most probably invested for different taxa and archipelagos, the species richness data used here are the most recent available.
Multiple regressions
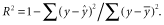
We used these R2 values to quantify the proportion of the total variation among archipelagos in species (indigenous, natives and endemics) explained (accounted for) by selected LMMs. If more than one model was selected as ‘best’ then we estimated for each variable the weight of evidence, i.e. the sum of AICc weights (wAICc) derived from the AICc for the models in which the variable is included, which measures the relative importance of each variable (Burnham & Anderson, 2002; see also Cameron et al., 2013). The variables with a wAICc = 1 are included in all best models. For the number of islands in each archipelago, we ran analyses for current island number and after considering palaeo-connections separately.
Endemism
Similar proportions of endemics among taxa could lead to converging results between indigenous and endemic species. We tested whether the proportion of endemics differs among snails, birds and plants (for spiders, data are available for endemics only). The proportions differ statistically [Kruskal–Wallis test: H (d.f. = 2, n = 42) = 12.3, P = 0.002)]. Removing birds from the three Atlantic archipelagos did not alter this result [Kruskal–Wallis test: H (d.f. = 2, n = 39) = 12.8, P = 0.002)].
Model-based inference of biogeographic rate scaling
After determining which predictor variables correlate with species richness, we performed additional analyses to investigate which biogeographic rates, i.e. cladogenesis, anagenesis, extinction and colonization, are likely to be affected by those variables. Due to the complexities related to island age, and to simplify the analysis, we retained only the three biogeographic variables with the maximum importance values in the multiple regressions: area, isolation and number of islands (see Results, Table 2). We sought to determine whether each variable influenced species numbers through its effects on the scaling of colonization, extinction or speciation rates. Furthermore, we asked whether similarities in the scaling of species richness with predictor variables across taxonomic groups were reflected by similarities in the scaling of biogeographic rates.
| Indigenous | Natives | Endemics | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model 1 | Model 2 | Model 3 | Variable importance | Model 1 | Model 2 | Model 3 | Model 4 | Variable importance | Model 1 | Model 2 | Model 3 | Variable importance | |
| Intercept plants | 1.57 (0.16) | 1.36 (0.15) | 1.76 (0.26) | 1 | 5.13 (0.58) | 4.43 (0.48) | 3.865 (0.57) | 4.59 (0.72) | 1 | 0.59 (0.23) | 0.77 (0.28) | 0.63 (0.54) | 1 |
| Contrast birds | −1.24 (0.04) | −1.238 (0.04) | −1.24 (0.04) | 1 | −1.36 (0.08) | −1.36 (0.08) | −1.36 (0.08) | −1.36 (0.08) | 1 | −1.21 (0.09) | −1.214 (0.09) | −1.88 (0.56) | 1 |
| Contrast snails | −0.66 (0.04) | −0.66 (0.04) | −0.66 (0.04) | 1 | −1.19 (0.08) | −1.19 (0.08) | −1.19 (0.08) | −1.19 (0.08) | 1 | −0.36 (0.09) | −0.36 (0.09) | −0.59 (0.56) | 1 |
| Contrast spiders | – | – | – | – | – | – | – | – | – | −0.63 (0.09) | −0.63 (0.09) | 0.55 (0.56) | 1 |
| Area total | 0.57 (0.05) | 0.60 (0.05) | 0.57 (0.05) | 1 | – | – | 0.147 (0.09) | 0.11 (0.09) | 0.37 | 0.73 (0.08) | 0.71 (0.08) | 0.73 (0.08) | 1 |
| No. of islands | −0.91 (0.15) | −0.85 (0.17) | −0.75 (0.16) | 1 | – | – | – | – | 0 | −0.97 (0.26) | −1.02 (0.25) | −0.94 (0.27) | 1 |
| Isolation | – | – | −0.13 (0.07) | 0.27 | −0.81 (0.16) | −0.69 (0.16) | −0.66 (0.15) | −0.76 (0.16) | 1 | – | – | −0.01 (0.17)* | 0.22 |
| Age | – | – | – | 0 | −0.39 (0.22) | – | – | −0.32 (0.21) | 0.461 | – | – | – | |
| Latitude | −0.01 (0.002) | – | – | 0.44 | – | – | – | – | 0 | – | −0.004 (0.004) | – | 0.23 |
| R2 | 0.97 | 0.97 | 0.97 | 0.94 | 0.944 | 0.943 | 0.94 | 0.86 | 0.86 | 0.90 | |||
| No. parameters | 8 | 7 | 8 | 7 | 6 | 7 | 8 | 8 | 9 | 12 | |||
| ΔAICc | 0 | 0.83 | 0.94 | 0 | 0.09 | 0.67 | 1.68 | 0 | 1.72 | 1.80 | |||
| wAICc | 0.44 | 0.29 | 0.27 | 0.32 | 0.309 | 0.230 | 0.140 | 0.547 | 0.23 | 0.22 | |||
- The most parsimonious are all models (e.g. Model 1, 2, etc.) with an AICc difference (ΔAICc) less than 2 and are the ones presented (see also Materials and Methods). Effect sizes (estimates of regression coefficients) are presented along with their standard errors (in parentheses). For the levels of the factor ‘taxon’, effect sizes for birds, snails and spiders are shown in the form of differences (contrasts) with respect to the absolute value for plants. The R2 value, the number of parameters in the model, the AICc difference and the AICc weight (wAICc) are given for each model. The importance of each variable, i.e. the sum of wAICc of models including them, is also given. Results for birds, snails and plants are for 14 archipelagos and for endemic spiders for 12. Results for birds are with the three Atlantic archipelagos, i.e. the Azores, Madeira and the Canary Islands, but their exclusion gives identical results (see Table S3 in Appendix S3). See also Table S4 in Appendix S3 for the same analysis but with the extant number of islands for each archipelago instead of paleo-connections.
- *Indicates taxon-variable interactions.
We focused on the numbers of archipelagic endemic species and the number of native species (i.e. indigenous but not archipelagic endemic) as our response variables. The numbers of archipelagic endemics and natives can change through the following events (Fig. 1): (1) colonization from the outside (adding one non-endemic species); (2) anagenetic speciation (subtracting one non-endemic and adding one endemic species); (3) cladogenetic speciation (one endemic becomes two endemics); and (4) extinction (subtracting a species from either category). For our main analysis, we assumed that non-endemic species cannot undergo cladogenetic speciation. However, we also considered the alternative assumption, that non-endemics could undergo cladogenetic speciation, and compared the results (see Fig. S5 in Appendix S1). Because we focus at the archipelago level, dispersal from one island to the other does not affect archipelagic species richness directly, and so we did not consider intra-archipelago dispersal in our model.

Conceptual diagram showing the basic rates controlling endemic richness and non-endemic native richness. In the model-based analysis, the rates (arrows) are allowed to be functions of predictor variables recovered as significant in the regression analysis.
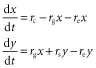
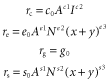
An exponent of zero corresponds to no effect of the predictor variable, > 0 and < 1 to sublinear scaling, 1 to linear scaling, and > 1 to superlinear scaling. The (x + y) term allows per-species speciation and extinction rates potentially to vary with total richness on the archipelago (i.e. diversity can be ‘regulated’). To facilitate our analysis, we assumed that archipelagic numbers of native and endemic species are at a steady state (dx/dt = dy/dt = 0). This can be enforced by assuming that the per-species extinction rate increases with total richness, and per-species cladogenesis decreases with increasing richness (e3 > 0, s3 < 0). The steady state is reached when inputs from colonization plus cladogenetic speciation balance the losses due to extinction, i.e. equilibrium (MacArthur & Wilson, 1967; but see Heaney, 2000). The possibility that per-species extinction rates might increase as richness increases is a commonly held view going back to original work on the theory of island biogeography (e.g. MacArthur & Wilson, 1967), and the diversity dependence of speciation rates has been supported by numerous analyses consistent with diversity dependence of speciation in phylogenetic studies (e.g. Phillimore & Price, 2008; see also Rabosky, 2009, 2013; Valente et al., 2014). Even so, the strength of the regulation of both speciation and extinction rates can freely vary in the analysis and, if justified by the data, one rate might show strong regulation while the other does not.
We used the deterministic rate of change equations to find the expected steady-state richness values, and assumed that deviations from the steady state of the log-transformed species numbers are Gaussian. This is similar to the error model used in most regression analyses of species–area curves. An alternative would be to allow the biogeographic rate parameters to determine the fluctuations around the steady state in a fully stochastic model (e.g. as a continuous-time Markov chain). We ultimately decided against the latter for reasons of computational efficiency, and furthermore it is not clear that all the deviations from expected values should be endogenous and linked to the rate parameters (i.e. exogenous factors such as biogeographic history or variation in sampling and taxonomic treatment could account for deviations from expected richness).
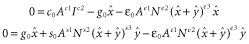
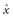 and
and 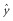 . The likelihood of a given parameter set, given vectors of observed species numbers (x, y), is then:
. The likelihood of a given parameter set, given vectors of observed species numbers (x, y), is then:

Bayesian parameter inference
We took a Bayesian approach to characterize the range of parameters that fit the data well. Given the limitations of the size of the dataset relative to the number of parameters in even a simple process-based island biogeography model, we aimed to assess the identifiability of the model parameters and evaluate which rates can be constrained by the data and which cannot. Species richness scaling might be more sensitive to colonization scaling than to speciation scaling, for example, and thus a particular richness scaling can constrain only those parameters. One cannot estimate the scaling of all rates with all potential predictor variables without additional data types (e.g. phylogenies for all taxa on all archipelagos) that are not available at the present time. A main challenge in this kind of analysis is overfitting: the number of parameters explodes when all effects are allowed to vary. We constrained intercepts to vary between 10−15 and 1010 and exponents to vary between −2 and 2. The only exception was the scaling of speciation with species richness, which is zero or negative (–2, 0) and the scaling of extinction with species richness, which is zero or positive (0, 2). The prior is a non-informative uniform distribution over the parameter space.
We used a Markov chain Monte Carlo (MCMC) algorithm to sample parameter combinations from the posterior. We used the ‘mcmcrun()’ function in the MCMC Toolbox in Matlab (Haario et al., 2006) to implement the sampling (code available from EPE). For each analysis, we ran 10 independent MCMC runs, and combined the post-burn-in posterior samples of all runs to give the final posterior. Burn-in was evaluated for each run individually by examining the stationarity of model parameters and consistency of sampled parameter estimates across runs. Overall convergence was assessed using consistency of posterior distributions between runs.
RESULTS
Multiple regressions – linear mixed effect models
According to the AICc-based model selection procedure, more than a single parsimonious model was selected for indigenous, native and endemic species. Considering only the variables with wAICc = 1, i.e. the variables included in all the best models and thus having maximum relative importance (i.e. empirical support; Burnham & Anderson, 2002), we selected total area (positive effect) and number of islands (negative effect) for indigenous and endemic species. For native species, isolation has a negative effect (Table 2). Latitude, isolation and the geological age of the archipelagos had lower, or no, importance across models (Table 2). The models explained most of the variation in species richness for the three distributional categories (R2 ≥ 0.92 for all cases). Total area and number of islands were not significantly correlated (P > 0.10).
No taxon × variable interactions were retained in the most parsimonious models, apart for isolation in a single case (see Table 2), indicating indistinguishable slopes across taxa for the relationships between species richness, total area and number of islands for indigenous and endemic species. All the best models for indigenous, native and endemic species richness revealed significantly different intercepts for all taxa (Table 2).
The indistinguishable slopes indicate that indigenous species richness scales to the same power with total archipelagic island area for three different taxa across archipelagos scattered across the globe (Fig. 2). The same is true for endemic species, and also regardless of the number of islands considered, i.e. taking into account palaeo-connections or using number of extant islands (Appendix S3). The slopes of the species–total area relationship in logarithmic space were 0.57 ± 0.04 SE for indigenous species and 0.72 ± 0.06 SE for endemic species, both of which are within the range of values proposed for the inter-provincial species–area relationship for continental landmasses (Rosenzweig, 1995) and higher than the typical slopes for the classic species–area relationship across individual islands (Rosenzweig, 1995; Ricklefs & Bermingham, 2007; Triantis et al., 2012). In contrast to the homogeneous slopes, the intercepts of the indigenous and endemic species–area relationships differed considerably among the taxa (Fig. 2, Table 2). Considering extant islands for each archipelago (Table S1 in appendix S1), without taking into account past connections of islands, gave similar results (Table S4 in Appendix S3).
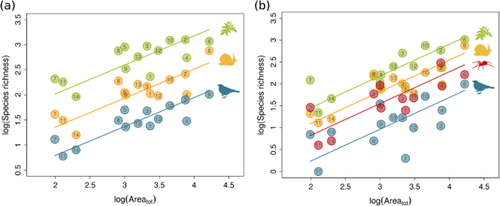
Individual relationships between archipelagic total area and taxon richness. (a) Indigenous and (b) archipelagic endemic species for land birds ( ), vascular plants (
), vascular plants ( ), land snails (
), land snails ( ) and spiders (
) and spiders ( ). The lines are the predictions from mixed effect models (Table 2), the effects of variables other than total area and interactions are included, using the mean value across archipelagos and wAICc-averaged coefficients. The slopes for each taxon are parallel (see Table 2). For birds we include the three main Atlantic archipelagos, i.e. the Azores, Madeira and the Canary Islands; however their exclusion provides similar results (Table S3, Fig. S3). Archipelagos are: (1) Comoros, (2) Galápagos, (3) Hawaii, (4) Juan Fernández, (5) Mariana Northern, (6) Marquesas, (7) Mascarenes, (8) Samoa, (9) Gulf of Guinea Islands, (10) Society Islands, (11) Tristan da Cunha, (12) Azores, (13) Canary Islands, (14) Madeira.
). The lines are the predictions from mixed effect models (Table 2), the effects of variables other than total area and interactions are included, using the mean value across archipelagos and wAICc-averaged coefficients. The slopes for each taxon are parallel (see Table 2). For birds we include the three main Atlantic archipelagos, i.e. the Azores, Madeira and the Canary Islands; however their exclusion provides similar results (Table S3, Fig. S3). Archipelagos are: (1) Comoros, (2) Galápagos, (3) Hawaii, (4) Juan Fernández, (5) Mariana Northern, (6) Marquesas, (7) Mascarenes, (8) Samoa, (9) Gulf of Guinea Islands, (10) Society Islands, (11) Tristan da Cunha, (12) Azores, (13) Canary Islands, (14) Madeira.
To test the absence of slope differences among taxa for the relationships between species richness and total area and number of islands, we also used average area, which summarizes the positive effect of total area and the negative effect of the number of islands. The results remained unchanged (Fig. S4 in Appendix S1, Table S5 in Appendix S3).
Model-based inference
In the independent models for each taxon, the posterior distributions generally agreed across taxonomic groups and with the model combining all taxa (Fig. 3), with exponents of similar sign and magnitude (or similar levels of uncertainty across the parameter space). To determine that the models produced reasonable results, we compared the model predictions from 1000 randomly chosen parameter sets from the posterior distributions with the empirical values for native and endemic richness, and they agreed well (Fig. S6 in Appendix S1) and also matched the empirical exponents of the richness–area scaling (Fig. S7 in Appendix S1).
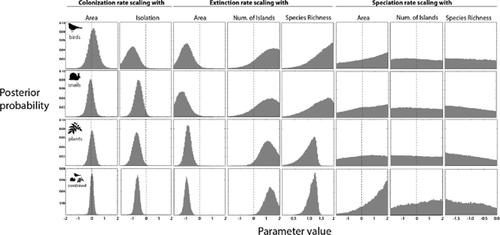
Sampled posterior distributions of scaling exponent parameters of different biogeographic rates and predictor variables. These are the posterior distributions of each parameter across the whole parameter space, not at a fixed values of other parameters. Each parameter reflects the scaling relationship between one rate (e.g. extinction) and one predictor variable (e.g. area). The top three rows reflect models fitting of each taxonomic group individually. The bottom row assumes common exponent parameters across taxonomic groups, but allows group-specific intercepts.
The combined model was generally the most informative with regard to conclusions about colonization and extinction, with the speciation parameters being less identifiable (Fig. 3). Colonization exhibits a negative relationship with isolation for all taxonomic groups. The extinction rate shows a negative relationship with area and a positive scaling with number of islands. Moreover, results for extinction provide support for diversity dependence, with extinction rates showing a positive relationship with species richness. The posterior distribution for in situ cladogenetic speciation rate, for which we did not consider isolation, is generally flat, indicating that the data are consistent with a range of cladogenetic speciation scalings and the parameters are not identifiable with these data, with the exception that for the combined data there seems to be (weak) evidence that the speciation rate scales positively with area. To test that our conclusions about colonization and extinction are not affected by uncertainty in the speciation parameters, we performed an additional analysis fixing a positive relationship of speciation with area, and found that the other inferred relationships were stable even given this constraint (see Fig. S8 in Appendix S1). The alternative model, in which endemics and non-endemics can undergo cladogenetic speciation, produced similar results (see Fig. S5 in Appendix S1), with slightly more evidence in the combined model that speciation rates are also dependent on the number of islands and area.
Discussion
The strikingly high proportion of variation in species richness that is explained by area and number of islands and the consistent scaling of species richness with area across taxa are puzzling. Within isolated archipelagos, diversity is mainly regulated by loss of species and the production of new species. These processes depend on the dynamic nature of oceanic archipelagos, expressed by the continuous change in number and size of islands, as well as attributes of organisms, including body size, generation time and dispersal ability (Ricklefs & Bermingham, 2007; Whittaker et al., 2008; Pigolotti & Cencini, 2009; Kisel & Barraclough, 2010). Colonization from outside typically plays a secondary role (e.g. Whittaker & Fernández-Palacios, 2007). Moreover, within- and between-island mechanisms of speciation generally operate only above certain thresholds of island size and archipelago configuration, which differ among taxa according to dispersal abilities and which typically produce change in the slope of the species–area relationship (Rosenzweig, 1995; Losos & Schluter, 2000; Triantis et al., 2008). Accordingly, we expected the relationships between species richness and area of the four taxa to have different slopes owing to their disparate ecological requirements, dispersal abilities, typical population sizes, average spatial extent of intraspecific gene flow and propensity to form new species (e.g. related to generation time; see Patiño et al., (2014) for single islands). However, even when the effects of colonization from outside archipelagos are removed by considering only endemic species, the slope of the species–area relationship increases, but remains homogeneous, for all the taxa (Fig. 2, Table 2). Additionally, the varying degrees of completeness of the archipelagos’ faunas and the fossil records (e.g. Cameron et al., 2013), and the possible inconsistencies among taxonomists working across widely scattered oceanic archipelagos, add to the underlying stochasticity and thus further support the robustness of our findings.
Emergent ecological correlates of species diversity might result from different combinations of underlying evolutionary mechanisms (Rahbek, 2005). For example, the positive scaling of species richness with area could reflect several independent mechanisms, including a decrease in extinction rate and increases in speciation and colonization rates with area; but more certainly from several of these mechanisms, possibly acting with different strengths depending on spatial scale and/or a taxon's life-history traits (Rosenzweig, 1995; Triantis et al., 2012). Our process-based model provided support for empirically and theoretically established island patterns: colonization is strongly linked to isolation; cladogenetic speciation and extinction are both correlated with area (MacArthur & Wilson, 1967; Rosenzweig, 1995; Whittaker & Fernández-Palacios, 2007; Pigolotti & Cencini, 2009; Kisel et al., 2011; Rosindell & Phillimore, 2011; Cabral et al., 2014). The model also highlighted the strong dependency of extinction rate on archipelagic parameters, while dependences are less clear for speciation rate. While higher levels of archipelagic fragmentation might elevate diversification and allow more species to coexist within the same total area, LMMs showed a negative relationship of species richness with the number of islands (Table 2). Our process-based model provides support for the notion that fragmentation, at this scale, reduces the area of the component islands below a size that can maintain viable populations, thereby increasing extinction rates (e.g. Kisel et al., 2011), without an obvious negative effect on the generation of archipelagic endemics (Losos & Schluter, 2000). In general, however, speciation scaling may be difficult to infer with the current data, and so incorporation of phylogenetic information would greatly assist in inferring the drivers of speciation rates.
Obvious differences in the ecology of the taxa considered here suggest that different contributions of colonization, extinction and speciation (within-island and within-archipelago) to species richness for any particular archipelago were to be expected for each taxon, as indicated by the varying levels of endemism (see Results and Table 1). Highly mobile organisms such as birds do not undergo species formation on small islands, although they can form new species on archipelagos of small islands (e.g. Grant & Grant, 2008). For such taxa, the number of endemic species in an archipelago is rarely the sum of single-island endemic species, in contrast to highly sedentary species such as snails, that disperse infrequently between islands. According to our processed-based model, the posterior distributions of scaling exponents for the different biogeographic rates, i.e. colonization, extinction and speciation, across the three different taxonomic groups were mostly similar (Fig. 3). For example, the scaling exponent of extinction with area is similar for the three taxa considered, and so extinction presented the same relationship to area regardless of taxon, i.e. negative. Hence, despite the varying contribution of each of these processes to the species richness of each taxon, the processes exhibit similar scaling relationships with isolation, area and number of islands. Similar scaling of the underlying processes that influence species richness provides a strong explanation for the parallel scaling of species richness itself (see Model-based inference in Appendix S3 and Figs S6 & S7 in Appendix S1).
The homogeneous slopes for indigenous and endemic species imply that speciation and extinction rates for each taxon respond similarly to changes in archipelagic area. Differences between the taxa in the intercepts of the LMMs suggest differences in the capacity of island archipelagos to support species ecologically at unit area (Triantis et al., 2012), and these intercepts match the likely ecological space required by species of each of the taxa (i.e. plants< snails < spiders < birds) (Öckinger et al., 2010; Triantis et al., 2012; Fig. 2). Clearly, the results of our model are limited by the selection of the predictor variables and the sample size (although minimized by the use of LMMs, cf. Cabral et al., 2014; Patiño et al., 2014). The results could also be further supported, or challenged, by additional data that were not available in this study, such as the phylogenetic structure of communities, which provide information on the effect of area and other variables on the persistence times of island populations and rates of lineage splitting (e.g. Cadena et al., 2005). However, even without such information our results agree with existing island theory and provide insight into the striking pattern described herein.
The biotas of each of the oceanic archipelagos are independently derived from different continental faunas and floras, and within-archipelago diversification is independent across archipelagos (Whittaker & Fernández-Palacios, 2007; Gillespie & Clague, 2009). We found that these systems have independently converged on the same relationship between diversity and ecological space, expressed by the total archipelagic area and the way it is apportioned among islands. The generality of the diversity scaling described here suggests that it is rooted in fundamental limits to the processes establishing diversity (Hutchinson, 1959; Ricklefs, 2004; Phillimore & Price, 2008; Rabosky, 2009; Economo et al., 2015). This tightly constrained scaling of diversity, and its underlying processes, can be considered as an emergent property of the endogenous dynamics of oceanic archipelagos, explaining to a large extent the high variation in species richness obtained with the best multiple regression models for indigenous and endemic species (Table 2). The relative roles of evolutionary and ecological processes, as well as the geological histories of archipelagos, in producing consistent diversity–area outcomes for different taxa remain to be determined by additional data, including the phylogenetic structure of island biotas. A possible explanation for a common species–area scaling could be related to the idea of the existence of a (taxon-specific) theoretical maximum species diversity, i.e. carrying capacity (Brown et al., 2001; Whittaker et al., 2008; Rabosky, 2013), which canalizes the filling of available ecological space by diversity-dependent biogeographic processes (Phillimore & Price, 2008; Rabosky, 2013). However, our results presuppose that the taxa disperse well enough to colonize the archipelagos in the first place. Other groups, such as non-flying mammals and lizards, would probably not show similar diversity scaling on the same 14 archipelagos because they have failed to colonize many of the more isolated ones, for example Hawaii, the Azores and Tristan da Cunha (Gillespie & Clague, 2009).
Overall, the modelling framework presented here represents a novel direction in linking biodiversity data to biogeographic rates. However, as with any new inquiry, we should proceed with caution when interpreting results. First, our assumptions could be violated; for example, archipelagos may not be at steady-state richness (e.g. Whittaker & Fernández-Palacios, 2007). A future direction would be to compare a model based on an equilibrium assumption with predictions of a non-equilibrium model in a model selection framework. Second, the current implementation is highly computationally intensive, making the evaluation of its performance across the vast parameter space difficult, and there may be regions of parameter space that confound inference. The development of more computationally efficient, and ideally fully stochastic, implementations would be a promising direction for further work and an improvement upon the analyses presented here.
Our approach could be extended to allow a broader array of assumptions and predictor variables, but additional data and different types of data (e.g. phylogenies) would probably be needed to improve the power of the existing model and fit more complex models. The development of fully stochastic implementations would be a promising direction for further work, building upon the analyses described here. The potential of a process-based model is that it can, in principle, generate predictions for different biological patterns (e.g. richness, phylogenetic structure, population structure) that arise from the same underlying ecological and evolutionary dynamics. Key challenges to such a model, especially for islands, would be to consider the possible effect on speciation and extinction of a species richness carrying capacity of an island (e.g. Whittaker et al., 2008; Valente et al., 2014), untangle the different temporal scales on which colonization and speciation act, and take into consideration that isolated, large archipelagos may never reach an equilibrium but rather their state is described by a dynamic disequilibrium (e.g. Heaney, 2000). In this context, oceanic archipelagos, seen as distinct spatial and evolutionary units, present unique opportunities for synthetic analysis in biogeography and ecology.
Acknowledgements
This work was supported a Fundaçã para a Ciência e a Tecnologia (FCT) Fellowship (SFRH/BPD/44306/2008) to K.A.T. and partially by PTDC/BIA-BIC/119255/2010. F.G. was supported by the ‘Range Shift’ project (PTDC/AAC-AMB/098163/2008) from FCT, co-financed by the European Social Fund. R.E.R. acknowledges support from the Curators of the University of Missouri. E.P.E. acknowledges the support of NSF (DEB-1145989) and OIST. We thank J. Price, A. Boyer, J. M. Fernández-Palacios, P. A. V. Borges, H. Schaeffer, A. Oikonomou, Ch. Barboutis, S. Sfenthourakis, R. Preece, R. H. Cowie, J. C. Rando, M. Carine, P. Cardoso and A. F. Martins for assistance with the data; Jim Brown, Rosemary Gillespie, Larry Heaney and Michael Borregaard for discussions during the 6th International Biogeography Society Conference; R. J. Ladle, L. Heaney, B. C. Emerson, R. J. Whittaker and, especially, J. Chase and J. Brown for critical comments on previous versions of the manuscript. A. Phillimore, P. Weigelt, two anonymous referees and the editors provided extensive and insightful comments and suggestions.
References
Kostas Triantis has a long-term fascination with the biogeography of islands and also works on conservation biogeography and macroecology. He is currently working on diversity patterns of oceanic archipelagos.
Evan Economo has interests in documenting, understanding and conserving biodiversity at a variety of scales.
François Guilhaumon is interested in understanding the distribution of different aspects of terrestrial and marine diversity, with the goal of informing regional and global conservation efforts.
Robert Ricklefs has been interested since graduate school in the distribution of birds on islands, particularly in the West Indies, and is currently working on the host and geographic distribution of malarial parasites of island birds.




