Effects of bio-based residue amendments on greenhouse gas emission from agricultural soil are stronger than effects of soil type with different microbial community composition
Abstract
With the projected rise in the global human population, agriculture intensification and land-use conversion to arable fields is anticipated to meet the food and bio-energy demand to sustain a growing population. Moving towards a circular economy, agricultural intensification results in the increased re-investment of bio-based residues in agricultural soils, with consequences for microbially mediated greenhouse gas (GHG) emission, as well as other aspects of soil functioning. To date, systematic studies to address the impact of bio-based residue amendment on the GHG balance, including the soil microorganisms, and nutrient transformation in agricultural soils are scarce. Here, we assess the global warming potential (GWP) of in situ GHG (i.e., CO2, CH4, and N2O) fluxes after application of six bio-based residues with broad C : N ratios (5–521) in two agricultural soils (sandy loam and clay; representative of vast production areas in north-western Europe). We relate the GHG emission to the decomposability of the residues in a litter bag assay and determined the effects of residue input on crop (common wheat) growth after incubation. The shift in the bacterial community composition and abundance was monitored using IonTorrentTM sequencing and qPCR, respectively, by targeting the 16S rRNA gene. The decomposability of the residues, independent of C : N ratio, was proportional to the GWP derived from the GHG emitted. The soils harbored distinct bacterial communities, but responded similarly to the residue amendments, because both soils exhibited the highest mean GWP after addition of the same residues (sewage sludge, aquatic plant material, and compressed beet leaves). Our results question the extent of using the C : N ratio alone to predict residue-induced response in GHG emission. Taken together, we show that although soil properties strongly affect the bacterial community composition, microbially mediated GHG emission is residue dependent.
Introduction
The ongoing growth of the world population, its increasing demand for feed, food, and fibers, in combination with climate change necessitates a more sustainable circular economy, imposing a need to recycle residues from bio-based production chains into agricultural land. However, closing the loop in a circular economy may have unwanted side effects, such as increased greenhouse gas (GHG) production. Therefore, the consequences of bio-based residue input in agricultural lands have thus led to the need for a comprehensive organic amendment strategy to attenuate GHG emission, while maintaining the soil carbon storage capacity, quality, and fertility (Paustian et al., 2016). Previous work investigated the response of soil microorganisms to nutrient amendments through addition of specific residues in a particular soil type, or in relation to specific microbial guilds mediating greenhouse gas turnover in agricultural soils as case studies (e.g., Jia & Conrad, 2009; Bannert et al., 2011; Ho et al., 2011, 2013; Harter et al., 2014; Bastida et al., 2015; Pereire et al., 2015; Stempfhuber et al., 2016). Case studies are informative, but the nonuniformity of the methodologies used and variable experimental duration complicate cross-study comparisons (Pan et al., 2010; Shade et al., 2016). Rarely is the response of the soil microorganisms, as well as other aspects of soil functioning defined by a suite of parameters tested simultaneously and systematically.
Addition of organic matter in agricultural soils may alter the abundance, composition (community richness and evenness), and biotic interaction among soil microorganisms (Bannert et al., 2011; Ho et al., 2011; Wagg et al., 2014; Hartmann et al., 2015; Sengupta & Dick, 2015; Chen et al., 2016). For instance, residue input seemingly selects for specific members of the soil microbial community (Kuramae et al., 2011; Kuramae et al., 2012). Moreover, soil carbon turnover and its storage capacity are in part mediated by microbial activity where gaseous carbon dioxide and methane can be released into the atmosphere via mineralization of organic matter and associated microbial respiration. Gaseous carbon dioxide and methane release can be further stimulated by amendment of organic matter, resulting from the mineralization of newly added carbon and/or the existing carbon fraction, that is, priming effect (Fontaine et al., 2003; Li et al., 2013; Kim et al., 2014; Bastida et al., 2015; Ho et al., 2015a). Apart from organic matter decomposition, soil-inhabiting microorganisms catalyze a multitude of pivotal ecosystem processes in agricultural soils (e.g., methane oxidation: Ho et al., 2015b; biological nitrogen fixation: Herridge et al., 2008; crop disease suppression: Mendes et al., 2011). Considering that microorganisms are key to ecosystem processes (Wagg et al., 2014; Werling et al., 2014), changes in the microbial community and (a)biotic interactions may thus affect the soil microbial mediated processes, with consequences for GHG emission.
Recognizing the limitations of cross-study comparisons and that the effects of organic matter amendment is likely residue and soil type dependent (Cayuela et al., 2010; Paustian et al., 2016), we aim to determine the response of in situ GHG (i.e., CO2, N2O, CH4) emission to derive the global warming potential (GWP) of two representative agricultural soil types (sandy loam and clays soils) in north-western Europe after amendments with different bio-based residues. In general, it is assumed that the decomposition of a bio-based residue is dependent on its C : N ratio (Cayuela et al., 2010). However, most work on decomposition of residue materials varying in C : N ratios have not considered other effects, such as that of GHG emissions (apart from CO2) and the effects of the residues may have on crop growth. In order to determine whether GHG emission is dependent on the C : N ratio of bio-based residues, we applied six bio-based residues with a broad C : N coverage (5–521) to the soils. In parallel, a litter bag assay was performed to assess the decomposability of these residues and relate it to the CO2 flux (microbial respiration) and C : N ratio. Likewise, we anticipate higher N2O emission with the addition of residues with lower C : N ratios, indicative of less recalcitrant materials with high N content. Targeting the 16s rRNA gene, we determined the response of the soil bacterial community composition and abundance, respectively, via IonTorrentTM sequencing and quantitative PCR (qPCR) to the bio-based residue amendments. In addition, we account for crop growth (common wheat; Triticum aestivum) performance after incubation of the residue-amended soils over three months under controlled conditions. These efforts were intended to elaborate and assess the impact of bio-based residue amendments on soil functioning notably on the microbially mediated GHG emission and to recommend the optimal use of bio-based residues in agriculture.
Materials and methods
Site description, soil sampling, and residues
Soils were collected from agricultural fields from two research stations of Wageningen University and Research, the Netherlands. Vredepeel (51°32′32″N, 05°50′54″E) and Lelystad (52°31′20″N, 05°34′57″E) represent sandy loam and clay soils, respectively. These are typical agricultural soils for the Netherlands, as well as other temperate regions. These fields were planted with potatoes (Solanum tuberosum) and left fallow after harvest before sampling. Soil physicochemical properties have been determined before (Ho et al., 2015b). The upper 10 cm of the soils were collected in October 2013 from 1 m × 1 m plots at random and composited as a single bulk soil. The soil was air-dried at room temperature before being sieved to 2 mm. The residues included in this study were comprised of materials with a broad C : N ratio ranging from sewage sludge (5.5) > aquatic plant material (14.0) > commercial compost (15.3) > lignin-rich organic waste stream (17.1; designated ‘wood material’) > compressed sugar beet leaves (28.0) > paper pulp (518.3). With the exception of the paper pulp, these residues have been described before (Ho et al., 2015b). The paper pulp was sourced from SCA Hygiene Products SUAMEER BV (the Netherlands) to represent a residue with a high C : N ratio and is characterized by a total C and N of 399.12 ± 12.26 μg C mg dw sample−1 and 0.77 ± 0.09 μg N mg dw sample−1, respectively. The residues were air-dried at 30 °C, crushed, and sieved to 2 mm prior to use.
Experimental setup, gas flux, and nutrient analyses
Mesocosms comprising of 2.5 kg soil were set up in six replicate per soil and residue amendment as described in detail before (Ho et al., 2015b). Briefly, the residues (dry weight) were added into the soils at a rate of 20 ton ha−1 as is the typical range in agricultural practice (Diacono & Montemurro, 2010), and deionized water was added and maintained at 65% soil water retention capacity during the incubation. The mesocosms were incubated at 15 °C in the dark for approximately two months (56 days), during which, carbon dioxide, methane, and nitrous oxide flux was measured using an Innova 1412-5i Photoacoustic Infrared gas analyzer (lumaSense Technologies, Ballerup, Denmark) connected to an automated sampler equipped with a moisture trap (Innova 1309 multiplexer, lumaSense Technologies). Prior to the gas flux measurement, the mesocosm was placed in a gas tight chamber (diameter × height: 24 cm × 40 cm) for 30 min to equilibrate soil-atmosphere gas exchange. Gas flux rate was determined over an hour from at least four time intervals by linear regression (R2 >0.98). Following the gas flux measurement, soil was sampled using an auger (diameter × height: 3 cm × 10–12 cm), homogenized, and an aliquot of the soil was stored in the −20 °C freezer till DNA extraction. The remaining soil was stored in the 4 °C refrigerator for not more than 14 days to determine the soil physicochemical parameters. To determine biomass loss from the residues, a litter bag assay was performed in parallel using the same setup and incubation conditions with the exception that the residues were contained in a litter bag (0.4 mm mesh size) buried with soil. After incubation, the soils (control and residue-amended) were homogenized by hand in the same pot and planted with two seedlings of common wheat (Triticum aestivum), of which the seeds had been pregerminated in the greenhouse at ~22 °C. The crop was watered thrice weekly until harvest (100 days). After harvest, the aboveground crop biomass was determined after drying in the oven at 60 °C till constant weight (approximately three weeks).
Nutrient concentrations in the soil (NOx, NH4+, and PO43−) were determined in 1 m KCl (1 : 5 dilution) extract using a SEAL QuAAtro SFA autoanalyzer (Beun-de Ronde B.V. Abcoude, the Netherlands). The total carbon and nitrogen contents were determined from oven-dried (40 °C for 5 days) and sieved (0.4 mm) samples using the Flash EA1112 CN analyzer (Thermo Fisher Scientific, Bleiswijk, the Netherlands). Ergosterol content, used as a proxy for fungal abundance, was determined as described before using the alkaline extraction method (de Ridder-Duine et al., 2006). As reported before (Ho et al., 2015b), the mean soil pH (1 m KCl), organic matter content (% loss on ignition), and C/N ratio for the sandy loam soil were 5.38, 4.74, and 17.30, respectively, and for the clay soil were 7.64, 4.79, and 15.27, respectively.
DNA extraction and quantitative PCR (qPCR) assay
DNA was extracted from three randomly selected mesocosms out of the six replicate per treatment, soil type, and time using the PowerSoil®DNA Isolation Kit (MOBIO, Uden, the Netherlands) according to the manufacturer's instruction. The concentration and quality of the DNA extract was determined using a NanoDrop 2000C Spectrophotometer (ThermoScientific, the Netherlands). The DNA extract was stored in the −20 °C freezer till further molecular analyses. A qPCR assay targeting the total 16S rRNA gene copies (EUBAC assay) was performed with the Eub338f/Eub518r primer pair (Fierer et al., 2007) with primer concentrations, PCR thermal profile, and subsequent quality check of the amplicons as described in detail before (Ho et al., 2015b). Plasmid DNA extracted from a pure culture (Collimonas fungivorans strain TER331) was used to prepare the calibration curve. The qPCR was performed in duplicate for each DNA extract using a Rotor-Gene Q real-time PCR cycler (Qiagen, Venlo, the Netherlands).
16S rRNA gene amplicon sequencing and data processing
The V4 region of the 16S rRNA gene was amplified using the bacterial primer pair 515F/806R (Caporaso et al., 2011). The 5′-end of the forward primer consisted of the P1 key (underlined) and a two-bp linker GT (bold) (515-F: CCTCTCTATGGGCAGTCGGTGATGTGTGCCAGCMGCCGCGGTAA). The 5′-end of the reverse primer consisted of the A key (underlined), a 12-bp Golay barcode (Caporaso et al., 2012), and the CC linker(bold)(806-R: CCATCTCATCCCTGCGTGTCTCCGACTCAG-Golay-barcode-CCGGACTACHVGGGTWTCTAAT). The PCR reactions were performed with NEB Phusion High-Fidelity Polymerase (NEB Cat# M0530L). The PCR reactions were carried out on a Biorad iCycler (Bio-Rad Laboratories B.V., Veenendaal, the Netherlands) using the following program: initial cycle 98 °C for 30 s, followed by 25 cycles of 98 °C for 10 s, 50 °C for 30 s, and 72 °C for 15 s, followed by a final extension step at 72 °C for 7 min. Each PCR reaction (Total volume 50 μL) consisted of 0.5 μL of forward and reverse primers (5 μm of each primer), 10 μL of 5 × New England BioLabs Phusion High-Fidelity Polymerase reaction buffer, 1 μL of 10 mm DNTPs, 1.5 μL DMSO, 3 μL of template DNA, 0.5 μL Phusion High-Fidelity polymerase, and 33 μL nuclease free. The PCR products were purified with the AMPure XP PCR Purification Kit (Agencourt Cat# A638881) and subsequently quantified with Qubit Fluorometer (Thermo Fisher, Invitrogen, Bleiswijk, Netherlands). The samples were then pooled in equimolar ratios and subjected to IonTorrentTM sequencing.
The DNA fragments were sequenced unidirectionally using the IonTorrentTM semiconductor technology (Thermo Fisher Scientific) upon two batches of library prepared from 26 pM of equimolarly pooled samples for the sand and clay samples, respectively. The template was made by emulsion PCR using the Ion OneTouch™ 2 System with the Ion PGM™ Template OT2 400 Kit and subsequent enrichment for library containing ion sphere particles. The sequencing was performed using HiQ Ion PGM™ Sequencing 400 kit on Ion PGM™ System using two Ion 318™ Chips v2. For data processing, the forward and reverse primers were removed for each sequence from the sample FASTQ files using flexbar version 2.5 (Dodt et al., 2012). Sequences were trimmed by running in the Sickle tool (Joshi & Fass, 2011) to give a minimum and maximum read length of 25 and 150, respectively, before being converted into FASTA format and concatenated into a single file. Subsequently, all reads were clustered into OTUs employing the UPARSE strategy by dereplication and were sorted by abundance (with at least two sequences) and clustered using the UCLUST algorithm (Edgar, 2010). Next, chimeric sequences were detected using the UCHIME algorithm (Edgar et al., 2011). These steps were implemented in vsearch version 1.0.10 (Rognes et al., 2016), an open-source, 64-bit multithreaded compatible alternative to USEARCH. Prior to the dereplication step, all reads were mapped to OTUs using USEARCH global method to create an OTU table and converted to BIOM-Format 1.3.1 (McDonald et al., 2012). The sequences have been processed into the BIOM file using Hydra (de Hollander, 2017). Taxonomic information for each OTU was added to the BIOM file by aligning the sequences to the RDP database (release 11) (Cole et al., 2014) using SINA (Pruesse et al., 2007). These steps were implemented using snakemake version 3.1 (Köster & Rahmann, 2012). To determine the phylogenetic relationship between OTUs, the OTUs were first multisequence aligned against each other using mafft v7.040 (Katoh & Standley, 2013), and an approximately maximum-likelihood phylogenetic tree was generated using fasttree v2.1.7 (Price et al., 2010). The OTU table, phylogenetic tree, taxonomic information, and meta data were used in the multivariate statistical analyses in the context of environmental parameters. Nucleotide sequences found in this study were deposited at the European Molecular Biology Laboratory Sequence Read Archive under the study accession number PRJEB20909.
Statistical analysis
The principal coordinate analysis (PCoA), derived from the genus level assignment of OTUs, was performed in r version 2.15.1. (R Development Core Team, 2012) using the capscale function in the vegan package (Oksanen et al., 2015). The ordination was visualized considering the community data (OTUs at 3% divergence) using different distance measures: Bray–Curtis considers the species abundance count and Unweighted Unifrac considers the phylogenetic distance between the branch lengths of OTUs observed in the different samples without accounting for the abundances. The samples were grouped for different conditions, as well as the mean ordination value and spread of points (standard deviations of the weighted averages as eclipse using the Ordiellipse function in Vegan). Noneuclidean distances between objects and group centroids were handled by reducing the original distances (Bray–Curtis or Unweighted Unifrac) to principal coordinates and then, performing anova on these data. Analysis of variance using the distance matrices (Bray–Curtis/Unweighted Unifrac), that is, partitioning distance matrices among sources of variation (both quantitative and qualitative information) was performed using Adonis in vegan. This function, hitherto referred to as permanova, fits linear models to distance matrices and uses a permutation test with pseudo-F ratios.
The DESeqDataSetFromMatrix function in the deseq2 package (Love et al., 2014) was used to find OTUs/genera that were significantly distinct between different conditions with a significant cut-off value of 0.001. The DESeqDataSetFromMatrix function allows negative binomial GLM fitting (as abundance data from metagenomics sequencing is over-dispersed) and Wald statistics for abundance data. After performing these multiple testing corrections, OTUs/genera with log-fold changes between multiple conditions are reported. The same test was applied on the genera table which was obtained by binning the OTUs at genus level, with unresolved OTUs grouped as a single ‘unknown’ category. Next, the OTUs were visualized as boxplots after applying a log-relative transformation on the abundance table and given as Venn diagrams. The scripts and workflows for all the analysis above can be downloaded at: http://userweb.eng.gla.ac.uk/umer.ijaz#bioinformatics.
Level of significance between amendments and soils was determined in sigmaplot version 12.5 (Systat Software Inc., USA) using anova or t-test.
Results
Greenhouse gas fluxes
Residue amendments increased CO2 flux in both soils, with the exception of the compost- and wood material-amended soils where CO2 emission was comparable to the un-amended soils over the 56-day incubation (Fig. 1). In particular, cumulative CO2 emission appreciably increased after amendment with the aquatic plant material and compressed beet leaves. The spike in CO2 emission occurred at < 21 days; after 21 days, CO2 flux remained relatively constant till day 56 (Fig. 1).
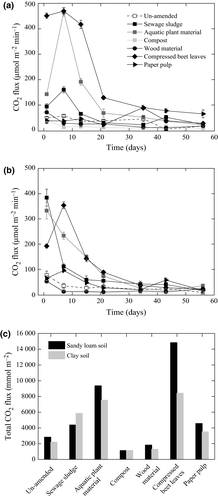
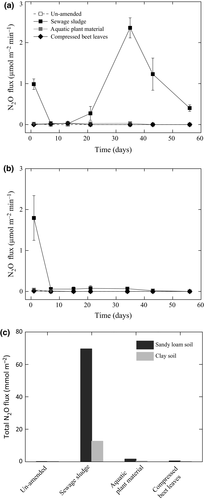
N2O emission appreciably increased after sewage sludge amendment in both soils. N2O emission also increased after the addition of aquatic plant material, but little or no N2O could be detected in the un-amended and other residue-amended soils (Fig. 2). The paper pulp had minimal or no effect on all the gas fluxes in both soils (Figs 1 and 2). It should be noted that the gas fluxes were determined in the absence of wheat and may exhibit different emission trends in the presence of the crop.
Litterbag assay, crop biomass, and soil nutrient status
The litterbag assay was performed to determine and relate residue decomposability through weight loss to CO2 flux; the CO2 flux was used as proxy for soil respiration. The residues with a relatively high weight loss in the litter bag were the sewage sludge (12–15%), aquatic plant material (40–45%), and compressed beet leaves (55–60%) (Fig. 3). This trend was consistent, showing comparable residue weight loss in both soils, except paper pulp, of which the loss was significantly higher (P < 0.05) in the clay soil (~16%) than in the sandy loam soil (~4%) (Fig. 3). Compost and wood material showed no or marginal weight loss at 0.5–1.3%, respectively.
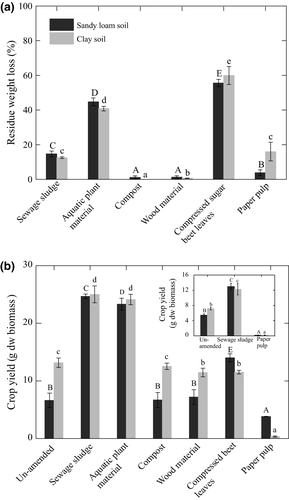
After incubation, wheat was planted in the pots to assess the effects of residue amendment on crop performance. Biomass yield, as determined by shoot biomass dry weight, was approximately 50% higher in the clay soil compared to the sandy loam soil (Fig. 3). Significantly higher crop biomass was recorded in the sewage sludge- and aquatic plant material-amended soils, whereas compressed beet leaves stimulated crop growth only in the sandy loam soil (Fig. 3). While crop yield in the compost- and wood material-amended soils was comparable to the un-amended soils, showing no net effect of these residues on the crop performance, paper pulp amendments significantly suppressed crop growth in both soils. Additionally, the mesocosm study was repeated using the same soils (sampled in 2015) and residues giving contrasting crop yields (i.e., sewage sludge and paper pulp); results confirmed the stimulatory and suppression effects of sewage sludge and paper pulp, respectively, on crop growth, although overall crop yield was lower (Fig. 3; inset figure).
As reported before (Ho et al., 2015b), NH4+ concentration was highest after amendment with sewage sludge, but levels gradually decreased, while NOx concentration increased over time in both soils. This indicated nitrification and is in agreement with the observed appreciable higher N2O emission after sewage sludge amendment in this study (Fig. 2). This trend was also detected in the aquatic plant material-amended soils, albeit to a lesser extent (Ho et al., 2015b). In other residue-amended soils including the paper pulp, the novel residue in this study, nutrient levels remained relatively constant during incubation.
Ergosterol concentration was determined after 21 and 56 days of incubation and was used as a proxy for fungal abundance. Initial ergosterol concentration in both un-amended soils was comparable (Table 1). Consistent in both soils, ergosterol concentration significantly increased after amendments with the residues, reaching appreciably higher concentrations in the aquatic plant material-, compressed beet leaves-, and paper pulp-amended soils (Table 1). In some of these soils (e.g., both aquatic plant material-amended soils), ergosterol concentration decreased from 21 to 56 days after incubation, while in others, ergosterol concentration increased (e.g., compressed beet leaves-amended sandy loam soil) or remained relatively constant (e.g., compressed beet leaves-, and paper pulp-amended clay soil). Although significantly higher, ergosterol concentration in the sewage sludge-, compost-, and wood oxidation material-amended soils was relatively low (Table 1).
| Treatment | Sandy loam soil | Clay soil | ||
|---|---|---|---|---|
| 21 days | 56 days | 21 days | 56 days | |
| Un-amended | 0.40 ± 0.04a | 0.45 ± 0.07a | 0.38 ± 0.09a | 0.44 ± 0.06a |
| Sewage sludge amended | 4.33 ± 1.35d | 3.48 ± 1.13d | 1.55 ± 0.59b | 1.10 ± 0.26c |
| Aquatic plant material amended | 10.26 ± 0.97e | 3.55 ± 1.01d | 7.01 ± 0.66d | 3.85 ± 0.95e |
| Compost amended | 0.64 ± 0.17b | 1.03 ± 0.19b | 0.64 ± 0.28a | 0.63 ± 0.03b |
| Wood oxidation material amended | 1.49 ± 0.47c | 1.92 ± 0.34c | 1.60 ± 0.65b | 1.84 ± 0.26d |
| Compressed beet leaves amended | 0.94 ± 0.13c | 5.56 ± 1.32e | 9.86 ± 2.78d | 9.07 ± 2.23 g |
| Paper pulp amended | 10.06 ± 1.91e | 2.53 ± 0.20d | 4.90 ± 0.88c | 5.57 ± 0.65f |
Soil bacterial 16S rRNA gene abundance and composition
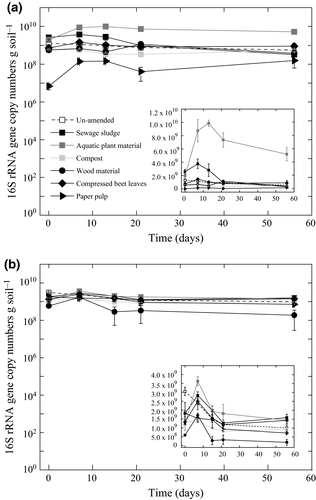
Previously, we characterized the methanotrophs quantitatively and qualitatively by targeting the pmoA gene (Ho et al., 2015b). Now, we focused on the total bacterial abundance and community composition by targeting the 16S rRNA gene. In the un-amended soils, 16S rRNA gene copies gradually decreased during the incubation, while in the sewage sludge-, aquatic plant material-, and compressed beet leaves-amended soils, the 16s rRNA gene copies increased, reaching a peak before values remained relatively constant or decreased (Fig. 4). This trend coincided with the spike in CO2 flux <21 days (Fig. 1). Although the 16S rRNA gene copies in the sewage sludge-, wood material-, and compressed beet leaves-amended soils were significantly altered after 56 days (pairwise comparison t-test; P < 0.05), values were not appreciably higher than in the un-amended soils. In the paper pulp-amended soils, mean 16S rRNA gene copies were lower than in the un-amended soil throughout the incubation.
The bacterial community composition was determined after 7 days (for both soils), and 13 days (for the clay soil), or 15 days (for the sandy loam soil), coinciding with the period with the highest CO2 emission assuming that the strong CO2 flux beyond background levels (un-amended soils) is caused by residue-induced microbial activity. The principal coordinate analysis (PCoA) revealed that the bacterial community composition was distinct in both soils, even after amendments with the same residues (Fig. 5). More pronounced in the sandy loam soil, the bacterial community composition was more similar following aquatic plant material and compressed beet leaves amendments than in the un-amended soils (Fig. 5). Indeed, we identified treatments (residue amendments), soil types, time (days), and pH to be highly significant (P = 0.001), respectively, accounting for 29.9%, 24.5%, 5.7%, and 3% of the variability influencing the community composition when we performed permanova of the community data against the different environmental parameters. Among the soil bacteria, identifiable predominant members fall into the Proteobacteria, Bacteroidetes, Actinobacteria, Acidobacteria, and Firmicutes at the phylum level, together forming the vast majority of the bacterial community composition (>70% relative abundance) in the sandy loam and clay soils (Fig. S1); differences in the community composition in these soils became apparent at the genus level (Figs S1–S3).
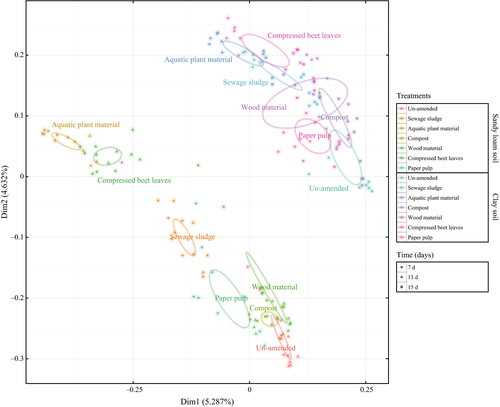
To detect discrepancies in the bacterial community composition between and within soils and treatments, we considered only OTUs showing strong changes (i.e., log-fold), termed as ‘significant’ OTUs/genera. Although the sequence length allows the identification of OTUs to the genus level, some OTUs could only be assigned to the family level (Fig. S1). Hence, the significant members of the community were represented here at the family level (Figs S2 and S3). After genus level assignment of the OTUs (boxplot; Figs S4 and S5), the significant genera between treatments were composed of community members belonging to the phylum Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, and Plantomycetes in both soils; undefined genera identified as incertae sedis in the Ribosomal Database Project (RDP) database were not considered (Figs S2 and S3). Besides, significant genera belonging to the Verrucomicrobia and Nitrospirae were only detected in the clay soil after sewage sludge and compost amendment, respectively. Unique to the sandy loam soil are members of Armatimonadates and Thaumarchaeota in the compost-amended soil and Acidobacteria in the beet leaves-amended soil.
Both soils harbored similar significant amounts of the members of the Bacteroidetes (i.e., families Flexibacteraceae, Sphingobacteriaceae, Chitinophagaceae, Flavobacteriaceae, and Cytophagaceae) although they do not necessarily occur in the same residue amendments in both soils (Figs S2 and S3). Actinobacteria was represented by some families (e.g., Conexibacteraceae, Glycomycetaceae, Promicromonosporaceae, Streptomycetaceae, and Acidimicrobiaceae) which were not common to both soils (Figs S2 and S3). Some Actinobacterial families only emerged after specific residue amendments (e.g., Acidimicrobiaceae in compost-amended clay soil, and all families in the residue-amended sandy loam soil, as no Actinobacterial genus was considered significant in the un-amended soil; Figs S2 and S3). Although Planococcaceae, Paenibacillaceae, and Bacillaceae belonging to Firmicutes are common in both soils, members of the family Turicibacteraceae, Leuconostocaceae, and Clostridiacea are unique significant genera detected in the wood oxidation material-, beet leaves-, and compost-/paper pulp-amended sandy loam soil, respectively (Fig. S2). Within Proteobacteria, significant genera fell into the subphylum Alpha-, Beta-, Delta-, and Gammaproteobacteria. Members of Gamma- and Deltaproteobacteria showed the highest variability without any shared significant genera between the soils and amendments (Figs S2 and S3). In contrast, with the exception of the wood oxidation material-amended clay soil, Oxalobacteraceae (subphylum Betaproteobacteria) was found to be significant in all residue-amended sandy loam and clay soils, including the un-amended clay soil (Figs S2 and S3). Within Alphaproteobacteria, Caulobacteraceae was significant in all residue-amended sandy loam soil, with the exception of the wood oxidation material-amended soil, but was not found to be significantly abundant in the clay soil (Figs S2 and S3).
Discussion
Response of microbial activity to bio-based residue amendment
The total CO2 emission trend was in agreement with the percentage residue loss as determined in the litter bag assay, in that, amendments with aquatic plant material and compressed beet leaves emitted the highest cumulative CO2 levels. This is in line with the documented highest residue weight loss in both soils (up to 60% during the 56-day incubation; Figs 1 and 3). In contrast, compost and wood oxidation material exhibited marginal weight loss in the litter bag assay (<2%) and also showed the lowest total CO2 emissions in both soils. Therefore, the aquatic plant material, compressed beet leaves, and sewage sludge can be regarded as readily degradable residues despite of their rather broad C : N ratios (5–28). The C : N ratio is typically used to assess the recalcitrance of organic matter, with a high C : N ratio (>18) indicating the presence of more complex organic material (Huang et al., 2004; Cayuela et al., 2010). Hence, our results question the extent of using C : N ratios alone to predict the soil response to residue input and consequences for greenhouse gas flux. While we cannot completely exclude a priming effect causing increased soil respiration following residue amendments (Fontaine et al., 2003; Cong et al., 2015), significant CO2 release coupled to a relatively higher biomass loss of the same residues suggest that the newly added carbon source, rather than the existing carbon fraction in the soil was mineralized, in which case, priming effect would be minimal.
As anticipated based on the relatively low C : N ratio (5.5), N2O emission was highest after sewage sludge addition. On the other hand, CH4 flux showed a more variable trend over time, depending on the residue, but independent of the residue C : N ratio (Ho et al., 2015b). We derived the global warming potential (GWP) in mg CO2 equivalent per kg soil (IPCC, 2007) by re-analyzing the cumulative CH4 flux (Ho et al., 2015b) in combination with the cumulative CO2 and N2O fluxes (Figs 1 and 2). The GWP value for CH4 and N2O is, respectively, considered to be 25 and 298 over a hundred-year time frame, while the GWP value for CO2 is regarded as 1 (IPCC, 2007). With the exception of the sewage sludge-amended sandy loam soil where N2O was the main contributor to the GWP, CO2 is the main contributor to the GWP for the other amendments. All residue amendments increased the mean GWP. Amendments with sewage sludge and compressed beet leaves, respectively, recorded a four- to ninefold and four- to fivefold GWP increase, whereas amendments with compost and wood oxidation material showed marginal GWP change (0.4- to 0.6-fold higher) when compared to the un-amended soil (Fig. 6). As the residues are used to replace or in conjunction with mineral fertilizers, future work could consider the GWP caused by amendments of a combination of residue and mineral fertilizers under field conditions.
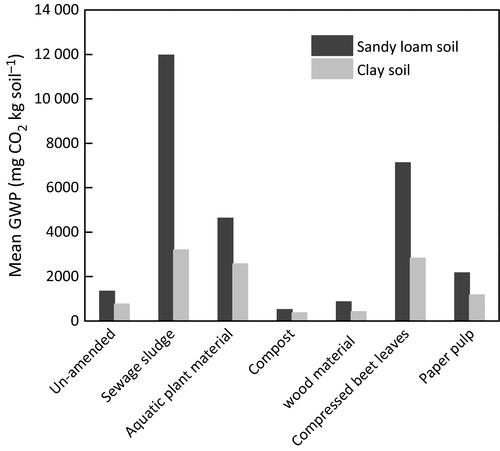
The CO2, CH4, and N2O emission patterns and rates responded similarly to the different residue additions despite the soils (sandy loam and clay soils) possessing different physicochemical parameters and soil texture (e.g., pH, C : N ratio; Ho et al., 2015b) and harbored distinct bacterial community composition (Fig. 5), indicating that residue input exerts a stronger impact on microbially mediated GHG emission than soil properties during the controlled incubation. Alternatively, although the bacterial community composition in both soils was distinct, they apparently shared similar traits, presumably as a result of a shared history (i.e., comparable agricultural regime), in mediating C- and N-cycling.
Trade-off between crop production and greenhouse gas emissions
In contrast to previous work solely determining greenhouse gas emissions and/or the response of the soil microbial community composition to (in)organic nutrient amendments (e.g., Huang et al., 2004; Ceja-Navarro et al., 2010; Serrano-Silva et al., 2011; Galvez et al., 2012; Odlare et al., 2012; Li et al., 2013; Sengupta & Dick, 2015), common wheat, a cereal grain crop, was planted in the mesocosms to assess crop growth and productivity (aboveground dry weight biomass) after residue amendments. Amendments with sewage sludge and aquatic plant material yielded higher crop biomass in both soils, in line with the higher N (NOx and NH4+) and P concentrations in the residue-amended soils prior to wheat addition (Ho et al., 2015b). This indicates nutrient limitation for crop, as well as for soil microorganisms. Using measured environmental parameters as constraints and qPCR data for a correspondence analysis revealed a positive correlation of NH4+ concentration and CO2 flux (Ho et al., 2015b), indicating a relief of N limitation for organic matter mineralization by soil microorganisms after residue input. Interestingly, paper pulp (in)directly suppressed crop growth. Repeating the experiment with the same soils (collected in 2015) and residues further confirmed our results (Fig. 3). The bacterial community composition in the paper pulp- and un-amended incubations was relatively similar as revealed by the principal coordinate analysis (Fig. 5), yet wheat biomass was significantly lower in the paper pulp-amended soils. Hence, the significantly adverse effect on plant growth is unlikely associated to the soil bacterial composition. Rather, the immobilization of available nitrogen caused by the high C:N ratio of paper pulp may lead to acute N-limiting condition for the crop. Therefore, not all bio-based residues are suitable soil additives. Although some residues may contribute to soil carbon (paper pulp C : N ratio: 518.3) and nutrients as in this study and others (e.g., Galvez et al., 2012; Kirkby et al., 2013; Pereire et al., 2015), the positive effects may be offset by detrimental effects on crop growth.
Among the residues tested, compost and wood oxidation material impose the lowest GWP, comparable to the un-amended soils. However, these residues did not yield higher crop biomass, indicating an apparent net zero effect with regard to overall greenhouse gas turnover (CO2, CH4, and N2O) and crop production, while causing minimal change in the bacterial community composition. Although seemingly having no net effect in the short term, the use of these residues is not without merit and may benefit other aspects of soil functioning. In particular, compost mineralization is slower than in fresh organic residues; hence, the extraneous carbon pool is sequestered in the soil for longer periods or is accumulated with recurring compost amendments (Ryals et al., 2015). Previously, we showed a significant stimulation of soil methane uptake with compost addition (Ho et al., 2015b). Although transient, compost-induced methane uptake offsets up to 16% of total carbon dioxide emitted during the incubation (Ho et al., 2015b). Therefore, agricultural soils may benefit from repeated compost amendments as means of a carbon sequestration strategy or by stimulating soil methane consumption, thereby reducing the GWP in agricultural lands.
Although crop yield was compromised in favor for reduced GWP, crop productivity, and properties may improve considering compost amendment complemented with other (in)organic soil additives (Ye et al., 2016). Application of inorganic fertilizer and organic residue mixtures is indeed common agricultural practice, which has also been shown to increase organic matter and nutrient retention (Kirkby et al., 2013; Chen et al., 2016). Hence, future studies under field conditions determining the optimum proportions of inorganic fertilizer and organic soil additives (i.e., to achieve optimum C : N : P ratio through combinations of these materials), as well as the application rate to minimize GWP while allowing sustainably high crop yields warrant further attention. More generally, our results reinforce the need for a more comprehensive approach when assessing ‘climate-smart’ agricultural soil management practices (Paustian et al., 2016; Ye et al., 2016).
Bacterial 16S rRNA gene copies and fungal abundances in residue-amended soils
Given that intensive Dutch agricultural soils are dominated by bacteria (Van der Wal et al., 2006), we followed the shift in the bacterial abundance over the incubation period and considered the community composition during the strong CO2 efflux. However, as certain fungi can respond rapidly to organic matter additions in agricultural soils (Van der Wal et al., 2013), the fungal biomass was determined after 21 and 56 days.
Despite a general trend showing the gradual decrease in the 16S rRNA gene copy numbers in the un-amended and residue-amended soils, values were higher (7–21 days) after reaching a peak at day 7 following sewage sludge and aquatic plant material amendments. The concomitant increase in the 16S rRNA gene copy numbers and abrupt CO2 increase (<21 days) beyond levels exhibited in the un-amended soils suggest residue-induced stimulation of bacterial abundance or growth (Figs 1 and 4). However, this trend was not observed in the compressed beet leaves-amended soils as would be anticipated if decomposition was bacteria mediated. Unexpectedly, the 16S rRNA gene copy numbers in the compressed beet leaves-amended soils remained relatively low and were comparable to values in the un-amended soils, albeit a significantly higher CO2 emission as well as a higher residue weight loss were detected. Instead, the ergosterol concentration in the compressed beet leaves-amended soils was appreciably higher than in the un-amended soils ≥ 21 days, indicating the relatively important role of fungal degradation in this amendment.
Ergosterol concentration was significantly higher (anova; P < 0.05; Table 1) in all residue-amended soils than in the controls, but was appreciably higher after aquatic plant material, compressed beet leaves, and paper pulp amendments in both soils. This suggests the relative dominance and stimulation of the fungal population after amendments with specific residues. Even in bacteria-dominated agricultural soils, fungi can respond quickly to addition of organic materials (Van der Wal et al., 2006). This appears to be the case in the current study. Therefore, fungi have contributed to the production of CO2. However, their possible involvement in the production or consumption of other GHG gases is not well-documented (Thomson et al., 2012).
Bacterial community response to bio-based residue amendments
The shift in the bacterial community composition following substrate addition had been documented before and could be related to the rapid CO2 efflux in soils (Fig. 1; Padmanabhan et al., 2003; Cleveland et al., 2007). Both soils harbored distinct bacterial communities, as revealed by the principal coordinate analysis with the plots depicting Bray–Curtis distances between the sandy loam and clay soils (Fig. 5). Phyla-level bacterial community response provides an overview, but may mask differential response among members of the same phylum (Ranjan et al., 2015, Morrissey et al., 2016; Ho et al., 2017). Therefore, we monitored the response of significant genera represented at the family level to bio-based residue amendments. The significant genera identified belonged to the phyla (e.g., Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria) common in agricultural soils from diverse geographical origins (Noll et al., 2005; Kuramae et al., 2012; Hartmann et al., 2015).
Interestingly, with the exception of the compressed beet leaves-amended sandy loam soil and sewage sludge-amended clay soil, members of Verrucomicrobia (families Opitutaceae and Verrucomicrobiaceae) and Acidobacteria (family Acidobacteriaceae) were not found to be significant in all other treatments, including the un-amended soils despite being shown to be ubiquitous and abundant across widespread agricultural and pristine environments (Jones et al., 2009; Bergmann et al., 2011; Kuramae et al., 2012; Ranjan et al., 2015; Kielak et al., 2016; Ho et al., 2017). The dominance of Opitutaceae and Verrucomicrobiaceae in the sewage sludge-amended clay soil thus suggests that these bacteria may have been introduced along with the residue and persisted in the soil, but their dominance was not observed in the sandy loam soil after sewage sludge addition. Although members of Verrucomicrobia and Acidobacteria are physiologically and taxonomically heterogeneous, studies consistently indicate that members of these phyla were more competitive under oligotrophic conditions (Fierer et al., 2007; Ho et al., 2017). In agreement with this presumption, bacteria assigned to the Verrucomicrobia and Acidobacteria appear to represent only a relatively minor fraction among the soil bacterial community after amendments, as well as in the un-amended soils. This suggests that the agricultural soils did not remain substrate/nutrient impoverished, as would be anticipated with ongoing agriculture where substrate/nutrient addition is a recurring practice.
Conversely, some bacterial families become important after residue input. These include Oxalobacteraceae of Betaproteobacteria and Chitinophagaceae of Bacteroidetes in the sandy loam soils after residue amendments (Fig. S2). In the clay soil, these bacteria also became significant, but only after amendments with certain residues (i.e., sewage sludge, aquatic plant material, and compost for Chitinophagaceae), as well as appearing in the un-amended incubations (i.e., Oxalobacteraceae) (Fig. S3). This suggests that members of Chitinophagaceae were more responsive to the residue amendments, and appreciably increased in relative abundance, more evident in the sandy loam soil. Hence, some members of particular phyla may indeed be disproportionately important to labile C-decomposition, but they are by no means definitely copiotrophs (please see review Ho et al., 2017). Tracking their persistence or cessation with labeled substrates and teasing apart other confounding factors (e.g., nutrient co-limitation) in prolonged incubation following the consumption of labile materials may reveal their ecological role.
The dominance of some bacterial families after residue amendments in the different soil types, but not in both soils after the same residue amendments, suggests that the soil properties override the effect of residue addition in altering the bacterial community composition. The PCoA supports this hypothesis, depicting a divergence in the relative abundance of taxons in both soils after residue amendments (Fig. 5) with some members contributing to the variability between the community compositions in these soils. For instance, significant genera constituting Planctomycetes appeared only in the clay soil, and those belonging to Gammaproteobacteria and Deltaproteobacteria are almost exclusive to either soils but not both (Figs S2 and S3). Nevertheless, the community composition after amendments with the more easily degradable residues (i.e., aquatic plant material, compressed beet leaves, and to a lesser extent sewage sludge) assuming higher respiration rate and residue weight loss as an indicator for degradability tended to cluster together (Fig. 5). This trend was more evident in the sandy loam soil where the bacterial community composition after amendments with the aquatic plant material and compressed beet leaves was more similar than in the un-amended soil. Consistent with previous work, soil properties (e.g., C : N ratio, pH) have been documented to be strong determinants and take precedence over land-use type and management in selecting for soil microorganisms (Lauber et al., 2008; Kuramae et al., 2012; Delmont et al., 2014), as well as their activity (Li et al., 2013).
Taken together, we conclude that soil properties are important determinants of the soil microbial community composition which take precedence over the effects of bio-based residue amendments. However, residue type, independent of C : N ratio, elicits a stronger response in the functioning of these communities with respect to GHG emission.
Acknowledgements
We are grateful to Iris Chardon, Marion Meima-Franke, and Tirza Andrea for excellent technical assistance. We extend our gratitude to Mattias de Hollander for assistance during the sequence analysis. UZI is supported by NERC Independent Research Fellowship NE/L011956/1. AH is financially supported by the BE-Basic Grant F03.001 (SURE/SUPPORT). This publication is publication nr. 6296 of the Netherlands Institute of Ecology.
Conflict of interest
The authors have no conflict of interests.




