Tracking lags in historical plant species’ shifts in relation to regional climate change
Abstract
Can species shift their distributions fast enough to track changes in climate? We used abundance data from the 1950s and the 2000s in Wisconsin to measure shifts in the distribution and abundance of 78 forest-understory plant species over the last half-century and compare these shifts to changes in climate. We estimated temporal shifts in the geographic distribution of each species using vectors to connect abundance-weighted centroids from the 1950s and 2000s. These shifts in distribution reflect colonization, extirpation, and changes in abundance within sites, separately quantified here. We then applied climate analog analyses to compute vectors representing the climate change that each species experienced. Species shifted mostly to the northwest (mean: 49 ± 29 km) primarily reflecting processes of colonization and changes in local abundance. Analog climates for these species shifted even further to the northwest, however, exceeding species’ shifts by an average of 90 ± 40 km. Most species thus failed to match recent rates of climate change. These lags decline in species that have colonized more sites and those with broader site occupancy, larger seed mass, and higher habitat fidelity. Thus, species’ traits appear to affect their responses to climate change, but relationships are weak. As climate change accelerates, these lags will likely increase, potentially threatening the persistence of species lacking the capacity to disperse to new sites or locally adapt. However, species with greater lags have not yet declined more in abundance. The extent of these threats will likely depend on how other drivers of ecological change and interactions among species affect their responses to climate change.
Introduction
Climate change is affecting ecological communities by shifting patterns of species’ phenology (Ellwood et al., 2013), elevational and latitudinal ranges (Lenoir et al., 2008; Amano et al., 2014), risks of local extinction (Parmesan & Yohe, 2003), and the nature of local adaptation and evolution (Aitken et al., 2008). Species’ migrations are expected to lag climate change given limitations associated with dispersal ability, generation time, and interactions with other taxa (Corlett & Westcott, 2013). While migration lags have been observed across a broad range of taxa (Bertrand et al., 2011; Chen et al., 2011; Devictor et al., 2012; La Sorte & Jetz, 2012; Schloss & Nuñez, 2012; Sunday et al., 2015), our understanding of the processes that affect the differential ability of species to track climate change remains incomplete. Most climate-impact studies to date have focused on assessing how species respond to large-scale shifts in climate, limiting our understanding of how species may differ in their responses to local shifts in climate (Dawson et al., 2011). Herbaceous communities may be particularly susceptible to climate change given that many species lack the capability to disperse long distances, limiting their ability to closely track shifts in climate (Van der Veken et al., 2007). However, shaded forest understories can also ameliorate local environmental conditions in ways that could delay impacts of climate change (De Frenne et al., 2013). Nevertheless, biological signals of climate change are still evident in the phenology of herbaceous plants, like those in central Wisconsin that now flower weeks earlier than they did earlier in the 20th century in apparent response to early-season warming (Bradley et al., 1999; Wright & Bradley, 2009; Ellwood et al., 2013).
Because the mechanisms by which plant species respond to climate change remain obscure, it is worth exploring whether species’ functional traits affect these responses. Functional traits reflect how organisms interact with their environment (McGill et al., 2006). We therefore expect them to affect how plant species respond to climatic factors and future climate change (Buckley & Kingsolver, 2012; Pollock et al., 2012). Traits vary in response to broad climate gradients (Wright et al., 2005) and affect local and regional processes of community assembly (Shipley et al., 2006; Xing et al., 2014). Fewer studies, however, use life history and functional traits to assess species’ responses to climate change. Life history traits and range characteristics served to predict extinction risk due to climate change in some regions (Pearson et al., 2014), but these responses are complex and rarely generalize to predict range shifts across a broad range of taxa (Angert et al., 2011). In studies resurveying multiple species across many sites, species’ traits can help to disentangle diverse drivers of ecological change (Leach & Givnish, 1996; Damschen et al., 2010; Crimmins et al., 2011; Soudzilovskaia et al., 2013; Amatangelo et al., 2014; Savage & Vellend, 2014). It is thus reasonable to suppose that traits might help explain the complex responses of individual species to climate change.
Site resurveys have identified marked changes in local and regional patterns of diversity and species’ incidence and abundance in the latter 20th century (Leach & Givnish, 1996; Damschen et al., 2010; Crimmins et al., 2011). The extensive ecological surveys made by J. T. Curtis and his students in the 1950s at more than 1000 sites (Curtis, 1959) provide a particularly useful baseline for evaluating the rate, nature, and extent of ecological change. Resurveys of forested sites in the 2000s (Waller et al., 2012) reveal frequent declines in local species richness (α diversity) and pervasive declines in compositional differences among sites (β diversity, reflecting ‘biotic homogenization,’ Rooney et al., 2004; Rogers et al., 2008). Upland forests in northern Wisconsin lost an average of 15% of native plant species, with conspicuous declines in taxa susceptible to deer herbivory and increases in exotic species and those avoiding or tolerating herbivory by deer (Rooney et al., 2004; Wiegmann & Waller, 2006). The upland forests of southern Wisconsin saw sharper declines in α and β diversity and more invasion by exotic species related to habitat fragmentation (Rogers et al., 2009). Sandy barrens dominated by open canopies of pines and oaks in the 1950s underwent succession in response to fire suppression, gaining some α diversity as they lost β diversity (Li & Waller, 2015).
Plant distributions in the upper Midwest are sensitive to climate as shown by the alignment of major ecotones along climatic gradients like the ‘tension zone’ in Wisconsin, Michigan, and Minnesota, where many species reach northern or southern range limits (Curtis, 1959). Cooler, moister, and snowier conditions prevail north of the tension zone. Over the past 50 years, winter and spring temperatures have increased, especially in northwestern Wisconsin (Kucharik et al., 2010). In summer, minimum temperatures have increased while drought and extreme precipitation events have become more frequent throughout the state. Climate models predict pronounced warming by midcentury, with increases in both the mean and variance of annual precipitation and accelerating rates of local climate change (WICCI, 2011; Ordonez et al., 2014).
Here we explore shifts in the distributions of 78 understory plant species over the past 50 years in Wisconsin and how these relate to shifts in local climatic conditions. We expect climate change to have affected plant distributions given the systematic climate changes that have occurred (WICCI, 2011). We hypothesize that shifts in species’ distributions have paralleled shifts in climatic conditions. However, we also appreciate that the ability of species to shift in distribution may be limited by various factors, including limited dispersal (especially in fragmented or heavily modified habitats), competition with other species (in which established plants have an inherent advantage), herbivory (e.g., by white-tailed deer), and other environmental factors. Using extensive resurvey data (Waller et al., 2012), we estimate shifts in species’ distributions using vectors connecting abundance-weighted centroids of the distribution of each species across 266 sites in Wisconsin surveyed in both the 1950s and 2000s. We further decompose these centroid shifts into vectors representing the underlying processes (local extirpation, colonization, and changes in abundance) and relate them to the centroid shift vectors to understand which processes structure species’ responses. To estimate how climates have shifted over this interval for each species, we apply climate analog analyses (Williams & Jackson, 2007; Veloz et al., 2011; Ordonez & Williams, 2013a). These allow us to compare the average climatic conditions species experienced in the 1950s to those experienced in the early 2000s by weighting the observed climate shifts by species’ abundances at each site. Using climate analog vectors for each species allows us to directly compare shifts in species’ distribution and abundance to the changes in climate that species has experienced. These comparisons reveal that shifts in species’ distributions have generally paralleled shifts in climate, but often fell short. We therefore ask whether variation in the magnitude of these lags is related to key life history and functional traits to learn more about the factors that may limit species’ responses.
Materials and methods
Field sampling
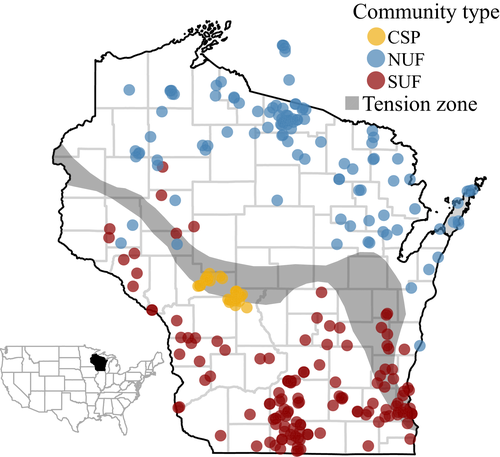
We analyzed changes across 266 study sites distributed across Wisconsin among three community types: southern upland forest (SUF), northern upland forest (NUF), and the central sand plains (CSP). These communities are compositionally distinct, with differing types and rates of ecological change over the last 50 years (Waller et al., 2012). Wisconsin Plant Ecology Laboratory staff surveyed the vegetation of Wisconsin at approximately 1000 sites between 1942 and 1958 to characterize community composition and species’ responses to environmental gradients (Curtis, 1959; Waller et al., 2012). They relied on plotless methods to characterize the overstory and sampled the understory using 20–50 spaced 1 m2 quadrats placed along a U-shaped or box-shaped transect. Within each quadrat, they noted all species present (herbs, shrubs, and tree seedlings), producing estimates of abundance based on their frequency across these 20–50 quadrats. Resurveys of the forested sites began in the 2000s using similar but more intensive sampling (80–120 quadrats) at 128 SUF sites (Rogers et al., 2008), 108 NUF sites (Rooney et al., 2004; Wiegmann & Waller, 2006), and 30 CSP sites (Li & Waller, 2015), for 266 sites in all (Fig. 1). Sampling design varied across community types (see citations above for specifics), but involved either replicating the original survey technique including more quadrats (SUF) or using a series of evenly spaced quadrats along parallel transects (CSP and NUF). Here we focus on more abundant species to ensure adequate sample sizes, including only native species found in at least 5 sites and 50 quadrats. This yielded an initial pool of 117 species.
Climate variables
We relied on 8-km gridded climate data of daily precipitation and minimum and maximum temperature between 1950 and 2006, spatially interpolated from an extensive network of weather stations to characterize historical climates (Kucharik et al., 2010). We computed seasonal summary variables from these data for each of our 266 sites using mean daily temperature and precipitation for fall, winter, spring, and summer; mean annual temperature and precipitation; annual temperature seasonality (coefficient of variation of mean monthly temperatures); and annual precipitation seasonality (standard deviation of mean monthly precipitation totals). We chose these summary variables to reflect a broad range of seasonal and annual aspects of climate change (Ordonez & Williams, 2013a).
Data analysis
Centroid shifts
Unlike many other studies that examine shifts in species’ ranges or distributions in terms of presence–absence data, we used abundance-weighted measures to depict species’ responses. These more accurately portray shifts in distribution, particularly in the short term as shifts in site incidence tend to lag behind shifts in climate (Chamberlain & Fuller, 2001; Virkkala & Lehikoinen, 2014). To measure shifts in geographic distribution, we calculated the abundance-weighted geographic centroid for each species in both the 1950s and 2000s. We weighted the mean latitude and longitude for each species at each site by its abundance (frequency) at that site, combining data across all sites. We then computed the bearing and distance for the shift in centroid for each species. These centroid shift vectors efficiently summarize changes in the distribution and abundance of each species across all the sites it occupied in the state. We then applied Rayleigh's test of uniformity to compare these bearings to a uniform circular distribution reflecting the null hypothesis of random centroid shifts. Rayleigh's statistic, r, quantifies the angular dispersion among the vectors from 0 (representing uniform dispersion) to 1 (indicating complete concentration in a single direction). We tested directionality of the centroid shifts by comparing the number of northward vs. southward shifts using a binomial test. We also compared the distances moved between these two groups using a two-sample t-test. Seven species were highly unusual in showing centroid shifts greater than 3 standard deviations beyond the shifts of other species. These outliers are so extreme that we considered it likely they reflected some artifact. To be conservative, we excluded them from further analysis leaving a pool of 110 species. In addition, our ability to track northward movements is restricted for species limited to northern Wisconsin as we could not track movements beyond its northern border. We therefore also excluded species whose Wisconsin distributional limits were confined to the northern two-thirds of the state (i.e., with a southern distributional limit above 43.5°). This further limited our species pool to 78 species, but reduced possible biases.
To assess how changes in overall abundance might affect the centroid shifts, we compared the distance and bearing of the centroid shifts to changes in abundance across and within community types. For each species, we calculated the regional proportional change in abundance over the last 50 years as the log of the ratio of total abundance across all sites in the 2000s divided by its corresponding abundance in the 1950s. We did the same for each species within each community type to understand whether the centroid shifts were being driven by losses or gains in particular habitats. These proportional changes in abundance were then used to predict centroid shift distances (via simple linear regression) and bearing (via circular regression). The circular regression model assumes the response variable has a von Mises distribution (Fisher & Lee, 1992), a circular analog of the Gaussian distribution (Lee, 2010).
Climate analog analyses
To estimate the climate shifts occurring at the sites where species occurred, we applied climate analog analyses. This approach generates output vectors comparable to the centroid shifts (Williams & Jackson, 2007; Veloz et al., 2011; Ordonez & Williams, 2013a). We determined spatial displacements in climatic conditions at each site by comparing its climate between 1950 and 1954 with the climates present at all grid cells in 2000–2004. We then calculated standardized Euclidean distances (SEDs) to each of the 2000s’ cells to identify which grid cell had the most similar climate (minimum SED). We then calculated the distance and bearing between each of the 1950s’ sites and its contemporary (2000s) climate analog cell.
To construct species-specific climate analogs, we then calculated a weighted average of the climate analog shifts for each species. As with calculating species’ centroid shifts, we summed the climate analog vectors across all sites that the species occupied in the 1950s, allowing us to calculate a weighted mean based on its original abundance there. The resulting vector provides a summary description of how climates have changed for each species across all occupied sites. We then compared these species-specific climate analog shifts to the corresponding centroid shift vectors for each species by calculating the difference between the two (hereafter referred to as the ‘lag’ vector) and exploring the correlation between these vectors of change. We also calculated the unweighted mean climate analog change vector across all sites. The abundance-weighted climate analogs provided a stronger signal for predicting shifts in species’ centroids than this overall vector of climate change (using dot products; see below for calculation). This suggests both that the climate signal is real and that species have responded individually to changes in climate. We acknowledge that our calculated lag vectors may not reflect the real migratory lag for each species. Species may be responding to very distinct sets of climate variables and their centroid vector may be highly dissimilar in direction from their climate analog vector. However, to facilitate comparison across species, we use this term regardless of whether species appear to be tracking climate change.
Vector correlation
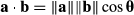
Here, ||a|| and ||b|| are the magnitudes of the species’ centroid and climate analog vector shifts, and θ is the angle between them. If the dot product is positive, the angle between the vectors is acute, while negative values reflect obtuse angles and zeros reflect orthogonal (independent) vectors. The dot product provides the mathematical basis of many correlation statistics, including the parametric Pearson's coefficient. Higher dot product values reflect stronger correlations between species’ centroid shifts and corresponding climate analog shifts, especially when these are manifest as larger displacements.
We tested statistical significance of the dot products using a randomization procedure. We randomized species’ abundances in the 2000s resurvey across all sites occupied during both time periods and calculated the resulting centroid shift and dot product. We repeated this procedure 1000 times to generate a distribution of dot products for each species and considered the observed dot product to be significant if it exceeded the 5th or 95th percentiles of the distribution. Species with significant negative dot products have centroids that are moving in a direction away from their vectors of climate change, while species with positive dot products indicate that shifts in species’ movements are in the same direction as the changes in climate those species experienced.
Decomposing the centroid shift vector
To better understand the processes underlying variation in species’ centroid shifts, we decomposed the centroid shift vectors into three components reflecting the colonization of new sites in the 2000s, local extirpation at the 1950s sites, and changes in abundance at the sites where species persisted over the study period. The origin of each vector was the species’ weighted centroid in the 1950s. For the colonization vector, the end point was an abundance-weighted centroid of all newly colonized sites in the 2000s. The local extirpation vector included the centroid for all sites where the species was lost over time. The change in abundance vector was an abundance-weighted centroid of all sites where the species persisted over the study period. As above, we used Rayleigh's Test of uniformity to test for the existence of a mean bearing. We additionally computed dot products between the centroid shifts and the vectors reflecting the underlying processes (colonization, local extirpation, and change in abundance). These allowed us to assess associations between these processes and overall shifts in species’ distribution. If species’ northward movement is driven by colonization and changes in abundance, we expect to find a positive correlation between the centroid vectors and the process vectors (i.e., a positive dot product). If the northward movement of centroid vectors is driven by local extirpation in southern sites, we expect to find the local extirpation vectors pointing in the opposite direction and a large proportion of negative dot products. Lastly, we used simple linear regression to relate the magnitudes of the process vectors and the lag vectors to estimate how the different processes predict species’ capacity to track climate change.
Trait analyses
We tested whether functional traits might account for differences among species in the extent of their centroid shifts and the degree to which their shifts lagged the observed climate analogs by measuring the association of these lags with 12 chemical, morphological, and physiological traits characterizing each species: seed dispersal mode, leaf carbon content, leaf circularity, leaf dry matter content, leaf length, leaf nitrogen content, leaf thickness, leaf width, seed mass, vegetative height, specific leaf area, and stem dry matter content. We measured each trait on at least 12 individuals (four individuals from each of three sites) following standardized protocols (Pérez-Harguindeguy et al., 2013). We assume that mean trait values have not changed since the 1950s and confirmed that trait variation is much greater among than within these species (D.M. Waller, unpublished data). These functional traits are known to affect species establishment, survival, reproduction, and/or leaf economics (Weiher et al., 1999; Westoby et al., 2002). Individual traits can display complex relationships when compared to a set of univariate climate variables (Moles et al., 2014). As we track climate changes using a multivariate representation of climate space, we cannot make specific predictions about how individual traits affect potential species’ responses to particular changes in climate.
We also compared species’ centroid shifts and lags to three other variables: each species’ initial area of occurrence (a convex hull drawn around the occupied sites in the 1950s), estimates of genome size for these taxa sampled in Wisconsin (Bai et al., 2012), and each species’ coefficient of conservatism (CC), an estimate of its habitat fidelity (Swink and Wilhelm, 1994). Values of CC range from 0 (no fidelity) to 10 (rarer species confined to specific, high-quality habitats). We expected that species with larger initial distributions would have more opportunities to respond to climate change through colonization/movement and therefore the lag between the movement of the species and its analog climate would be smaller. Larger genome sizes are associated with longer generation times and slower growth rates (Suda et al., 2014), suggesting that species with larger genomes might show greater climate lags. Species with narrower ecological habitat requirements (i.e., high coefficient of conservatism) might show larger lags and be more vulnerable to climate change if they are mostly restricted to small, dispersed patches of suitable habitat. For continuous traits, we used regression to assess how the magnitude of species’ centroid shifts and the lags between these shifts in centroids and climate analog vectors covaried with values of these traits. We applied anova to compare the magnitude of the lags among three different seed dispersal modes: unassisted, animal, and wind dispersal. We further evaluated the importance of dispersal traits using ancova to relate the magnitude of lags to seed mass, dispersal mode, and their interaction. We log-transformed traits that were highly skewed. Leaf dry matter content and stem dry mater content were highly correlated (Pearson's correlation coefficient = 0.85), as were leaf length and log leaf width (Pearson's correlation coefficient = 0.75). However, because we analyzed each trait separately, we included all traits in these analyses.
We used r (R Development Core Team) for all analyses, including the following packages: circular for circular regression (Agostinelli & Lund, 2013), geosphere for vector bearing and distance calculations (Hijmans et al., 2015b), ncdf4, raster, and dismo for processing climate variables (Pierce, 2014; Hijmans, 2015; Hijmans et al., 2015a), and ggplot2 for plotting (Wickham, 2009).
Results
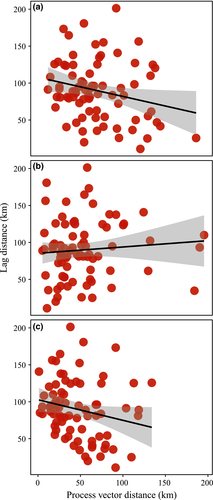
Over the 50-year study period, distributions of most common understory plant species in Wisconsin shifted substantially (Fig. 2a, Table S1). The centroid shift vector for a species efficiently summarizes both its initial mean distribution and its subsequent movement, aggregating quantitative abundance data across hundreds of sites. Species show considerable variation in the distance and direction of their geographic movement, but clear trends are evident. About 78% (61) of the 78 species display northward shifts, while 17 moved south (P < 0.001, binomial test). Northward shifts are also larger: 52.6 ± 30.3 km vs. 35.9 ± 20.6 km for southward shifts (means and SD's, Welch's t = 2.63, df = 38, P = 0.01). The average bearing of all centroid shifts was to the northwest (343°) and highly directional (P < 0.001 by Rayleigh's test). The overall strength of association among these vectors is intermediate (r = 0.54). The average distance of all centroid shifts is 48.9 ± 29.1 km. These centroid shifts were uncorrelated to the often conspicuous changes in species’ regional abundance we observed in these communities (Fig. S1, but species that gained abundance at the SUF sites tended to shift slightly more toward the northwest – Fig. S2f). These centroid vector shifts in distribution parallel higher rates of colonization and differential changes in local abundance for species persisting at those sites, with 63 and 70 of 78 species, respectively, having positive dot products. Local extirpation did not appear to drive these centroid shifts despite 46 of the 78 species having a negative dot product (Fig. S3). The colonization and change in abundance vectors both trend toward the northwest (with bearings of 352° and 338°, respectively) and are highly directional (P < 0.001 by Rayleigh's test for both). In contrast, the local extirpation vectors varied in magnitude and lacked consistent directionality (P > 0.05 by Rayleigh's test).
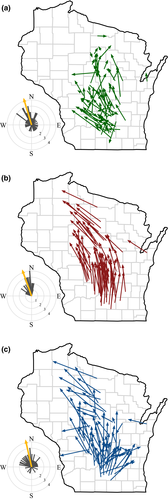
The climate analog analyses reveal that Wisconsin climates have changed considerably, but in heterogeneous ways over the past 50 years (Fig. S4). The climate analogs shifted an average of 114.3 ± 66.3 km, aggregated around a mean bearing of 343° (P < 0.001, Rayleigh's test). The species-specific shifts in analog climates are slightly shorter (99.7 ± 17.8 km), but more consistent in distance (compare SD's) and highly concentrated in direction (r = 0.93), clustered around a mean bearing of 340° (P < 0.001, Rayleigh's test) (Fig. 2b).
Shifts in species’ distributions over the past 50 years generally parallel the direction of shifts in analog climates (Fig. 2a, b). Three-quarters of the species (60 of 78) show a positive dot product between the species and climate-change vectors (mean = 0.19 ± 0.29), indicating an acute angle between the vectors (Table S1). Among these positive dot products, over half (32) show a significant dot product relative to the randomized expectation. Only 6 species had significantly negative dot products. None of the traits appear related to the magnitude of these species’ centroid shifts (Table S2).
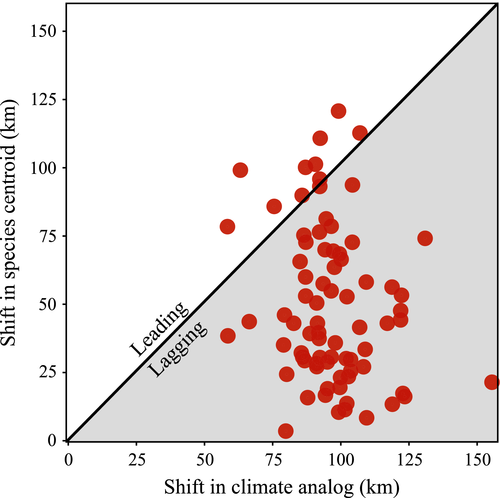
Although species’ distribution shifts thus appear to be tracking recent shifts in climate, most species’ centroid shifts seriously lag the estimated geographic shifts in climatic conditions (Figs. 2c and 3). Species’ centroid shifts average only 52% of the observed climate shift distances without accounting for direction. Taking into account the magnitudes and directions of both vectors, climate changes lead changes in species’ distributions by an average of 89.7 ± 40.4 km. Species’ centroid shifts usually track the direction of the climate analogs but cover only about 12% of the distance expected based on the changes in climate. The lag vectors representing the difference between the geographic shifts in climate and species’ centroid shifts are concentrated in direction (r = 0.60) with a mean bearing of 347° (P < 0.001, Rayleigh's test) (Fig. 2c). We find no relationship between overall changes in species’ regional abundance (log ratio of abundances) and the magnitude of the lags (linear regression: log ratio = 90.72 + 9.81*lag, F = 3.60, df = 76, P = 0.06, R2 = 0.04) as might be expected if some systematic driver (e.g., deer browsing) is acting to block species’ movements. However, species with greater seed mass, area of occupancy, and coefficient of conservatism all show slightly smaller lags (Table 1). None of these associations account for more than 10% of the variation in observed lags and no other continuous trait shows any relation to the lags. The magnitude of the lags did not differ among seed dispersal mode (anova: F(2,69) = 0.96, P = 0.39) or the interaction between dispersal mode and seed mass (Table S3). Species with higher magnitude colonization vectors showed smaller lags (Fig. 4a), although the relationship is weak and noisy. The other process vectors were statistically unrelated to the lags, however, the directions of the slope coefficients were in the predicted direction (Fig. 4b, c).
| Trait | Intercept | β estimate | SE (β) | t value | P value | R 2 | n |
|---|---|---|---|---|---|---|---|
| Area of occurrence | 90.05 | −9.64 | 4.54 | −2.12 | 0.04 | 0.06 | 77 |
| C-value | 87.84 | −7.66 | 4.86 | −1.58 | 0.12 | 0.04 | 63 |
| Coefficient of conservatism | 89.82 | −12.25 | 4.43 | −2.76 | 0.01 | 0.10 | 73 |
| Leaf carbon content (m2 g−1) | 88.80 | −7.41 | 4.55 | −1.63 | 0.11 | 0.04 | 71 |
| Leaf circularity (no units) | 89.36 | 0.06 | 4.71 | 0.01 | 0.99 | 0.00 | 72 |
| Leaf dry matter content (mg g−1) | 89.82 | −3.42 | 4.65 | −0.74 | 0.46 | 0.01 | 73 |
| Leaf length (cm) | 89.82 | −5.78 | 4.61 | −1.25 | 0.21 | 0.02 | 73 |
| Leaf nitrogen content (m2 g−1) | 88.80 | −5.06 | 4.60 | −1.10 | 0.27 | 0.02 | 71 |
| Log leaf thickness (mm) | 89.82 | −0.37 | 4.66 | −0.08 | 0.94 | 0.00 | 73 |
| Log leaf width (cm) | 89.82 | −7.45 | 4.58 | −1.63 | 0.11 | 0.04 | 73 |
| Log seed mass (g) | 91.21 | −10.33 | 4.47 | −2.31 | 0.02 | 0.07 | 70 |
| Log vegetative height (cm) | 89.82 | 0.45 | 4.66 | 0.10 | 0.92 | 0.00 | 73 |
| Specific leaf area (m2 kg−1) | 89.82 | 1.84 | 4.66 | 0.40 | 0.69 | 0.00 | 73 |
| Stem dry matter content (mg g−1) | 89.82 | −1.14 | 4.66 | −0.24 | 0.81 | 0.00 | 73 |
- Bolded values indicate statistical significance at an alpha level of 0.05.
Discussion
The unusually extensive baseline and contemporary data on plant abundances in Wisconsin allowed us to document 50-year changes in the distributions of 78 understory plant species and relate these to the corresponding shifts in climatic conditions observed across the state. Vectors reflecting shits in the distribution of these species are clustered and trend toward the northwest. The shifts in climate conditions we estimated for these species are also clustered and trend in the same direction, supporting the hypothesis that the shifts in plant distribution observed may reflect, at least in part, the changes in climatic conditions experienced by these species. These shifts in species’ distributions are correlated with the corresponding climate shifts, but differ considerably in the extent to which they match the magnitude of the climate shifts. Trends in both temperature and precipitation acted to drive the NW bearing of the climate analogs (Fig. S5) that often matched the direction of the species’ centroid shifts we observed. These results make clear that we should look beyond simplistic characterizations of climate change based only on temperature and general predictions that species will move up in latitude and elevation.
Climates have changed episodically throughout the quaternary with temperate plant species generally adapting by shifting their ranges in ways that allowed them to track these changes over broader spatial (subcontinental) and temporal scales (multiple centuries) (Davis & Shaw, 2001; Ordonez & Williams, 2013b). We observed that most of the species in this study seriously lag even the past half-century of climate change, albeit at finer spatiotemporal scales. Vectors of climate change outpaced those of species’ change by an average of 89.7 km, suggesting that over the last 50 years, most plant species have not closely tracked the climatic conditions that supported them 50 years ago. No native plant species went extinct in Wisconsin during the study period, but many have undergone considerable changes in regional abundance (Wiegmann & Waller, 2006; Rogers et al., 2008). We did not find overall changes in species’ abundance to be related to their respective lags, however, we did find that species’ ability to colonize new sites in the 2000s decreased the magnitude of the lags. Taken together, this suggests that while lags have not yet begun to substantially affect local species’ abundance and persistence, colonization of new sites since the original survey is critically related to tracking climate change. Overall, the ability of these plant species to keep pace with climate change appears limited and may become more so, particularly if changes in climate are accelerating and barriers to colonization increase.
Species’ lags were unrelated to extirpation and changes in abundance, further underscoring the difficulty in predicting population dynamics under future climate change and land use scenarios. Landscape context and habitat fragmentation have substantially influenced colonization events in the southern upland sites, but extirpation appears to be more stochastic (Rogers et al., 2009). Southern sites face an extinction debt wherein smaller patches of forest surrounded by urbanization are experiencing higher risks of extinction (Rogers et al., 2008, 2009). As climate change accelerates, these fragmentation effects could slow dispersal and thus the arrival of species better adapted to current climatic conditions. Species may also adapt to changing climates through phenotypic plasticity and/or local genetic adaptation. However, many understory species have long life spans, limiting opportunities for selection. If the cooler and moister conditions present under forest canopies act to buffer these habitats against warm and dry conditions (De Frenne et al., 2013), species may gain time to disperse and adapt. Thus, differences in dispersal, habitat occupancy, and other forces may cause extinction risks to vary, explaining the lack of consistent extirpation vectors related to distribution shifts in these species. McCune & Vellend (2015) also found 40-year extirpation events to vary greatly in ways unrelated to plant traits. With so many forces acting, extinction dynamics are likely to be complex and difficult to predict.
Climate changes vary geographically (Ackerly et al., 2010). Likewise, species’ shifts in apparent response to climate change are also spatially complex and individualistic (Grenouillet & Comte, 2014). Several factors appear related to the variation we observed in colonization and local changes in abundance among these species. Many species may lack the capacity (or opportunities) to disperse adequately to keep up with climate change, particularly in fragmented habitats (Pearson et al., 1998; Verheyen et al., 2004). Species that disperse ballistically, via ants (myrmecochory), and those lacking adaptations for dispersal may have difficulty colonizing new sites, although we do not find evidence here (anova comparing magnitude of colonization vectors among seed dispersal mode: F(2,69) = 2.74, P = 0.07). We also expected those with larger seeds and habitat specialists with conservative life histories to be inefficient at colonizing new sites (Estrada et al., 2015). We found that species with smaller areas of occupancy tended to lag shifts in analog climates more, as might be expected if they are dispersal-limited. However, we also found that species with larger seeds and higher coefficients of conservatism (habitat specialists) exhibited lower lags, counter to expectations. Although smaller seeds generally enhance mobility (Thomson et al., 2011), the dispersal of larger seeds could reflect fruit adapted for animal (bird or mammal) dispersal. Among our focal species, animal-dispersed species indeed had larger seeds on average (mean = 172 mg) than species with unassisted (24 mg) or wind-dispersed seeds (12 mg) (anova: F(2,66) = 5.65, P = 0.005, followed by Tukey's HSD test). Widespread logging in northern Wisconsin could also limit suitable sites for colonization or rates of dispersal for shade-adapted species. Because logging also acts to boost local deer densities, it could also increase deer impacts on species palatable to deer, lowering their density, reproductive success, and opportunities for local recruitment (Waller, 2014).
Few studies have examined how functional traits may mediate how plant species respond to climate change. Soudzilovskaia et al. (2013), however, noted in their study of alpine plants that species with larger seeds responded less to climate change, the opposite pattern from what we observed. It may be that environmental conditions and/or landscape context mediate how plants that differ in functional traits respond to changes in climate. In our study, the extent of climate change varied strongly with location, with higher rates of change in parts of northern Wisconsin. This may have created or increased the size of lags for species (and traits) particular to that area. Although functional trait studies have long examined how traits covary along environmental gradients, the literature on how traits affect changes in abundance is less developed. D. Li, A. Ives, D.M. Waller (in review) found several traits associated with patterns of species’ incidence among the CSP sites, but less related to differences in abundance. We only find weak evidence here of traits predicting geographic shifts over time, but this may reflect the spatial scale of the study and the fact that we aggregated abundance across sites rather than predicting changes within sites.
Several forces appear to be driving the changes observed in these Wisconsin plant communities over the past 50 years. Herbivory by white-tailed deer in northern Wisconsin has reduced the abundance of many understory species, including many broad-leaved herbs, while enhancing the abundance of graminoids and some ferns that better avoid or tolerate deer herbivory (Frerker et al., 2014). Many understory species also show declining rates of colonization, contributing to the species’ declines and biotic homogenization observed in these communities (Wiegmann & Waller, 2006; Rogers et al., 2009; Li & Waller, 2015). Yet despite these other strong drivers of ecological change, the results presented here strongly support the conclusion that changes in climate have already begun to alter the abundance and distribution of Wisconsin forest plants. The complex effects of these other drivers likely limit and modify how these plant species can respond to climate change, contributing to the complexity of patterns found and the apparent climate lags observed among these 78 species. We also note that these lags represent limited estimates of species’ responses to climate change given that we lack data on shifts in abundance and distribution beyond Wisconsin. Given these effects, the ability of species to track changes in climate within the state seems likely to affect the future composition of local communities and perhaps the floristics of the state as a whole.
Species’ distribution patterns reflect the effects of a complex mosaic of interacting biotic and abiotic factors (Sexton et al., 2009). Although we expect shifts in plant distribution to lag shifts in climate (Corlett & Westcott, 2013), these lags vary greatly in size, and probably importance, among species. Increases in the size of these lags could increase downward pressures on (or the intensity of natural selection within) populations that are already declining for other reasons (e.g., biotic interactions). Many species appear to lack the traits necessary to keep up with recent rates of climate change and other shifts in ecological conditions. As the rates of change in climatic and landscape conditions accelerate, these species will become more vulnerable and additional species will likely become vulnerable as well. Being able to quantify the gap between species’ current distributions and the climatic conditions they are adapted to will be of value for understanding the threats these species face and the ways diverse drivers of ecological change interact to threaten the persistence of many species.
Acknowledgements
The NSF Dimensions of Biodiversity Program provided essential funding for this work (DEB-1046355). C. Kucharik, D. Rogers, D. Li, G. Sonnier, and B. Alverson generously shared data for the analyses. We also thank P. de Frenne, S. Hotchkiss, A. Paulson, and J. Williams for closely reading an earlier version of the manuscript. The manuscript benefited greatly from comments provided by anonymous reviewers. DMW thanks I. Olivieri, O. Ronce, the LABex program, and ISEM for support during his sabbatical stay at the University of Montpellier, France.




