Incubation with porcine urinary bladder matrix yields a late-stage wound transcriptome in endothelial cells and keratinocytes isolated from both diabetic and non-diabetic subjects
Abstract
Proper wound closure requires the functional coordination of endothelial cells (ECs) and keratinocytes. In the late stages of wound healing, keratinocytes become activated and ECs promote the maturation of nascent blood vessels. In diabetes mellitus, decreased keratinocyte activation and impaired angiogenic action of ECs delay wound healing. Porcine urinary bladder matrix (UBM) improves the rate of wound healing, but the effect of exposure to UBM under diabetic conditions remains unclear. We hypothesized that keratinocytes and ECs isolated from both diabetic and non-diabetic donors would exhibit a similar transcriptome representative of the later stages of wound healing following incubation with UBM. Human keratinocytes and dermal ECs isolated from non-diabetic and diabetic donors were incubated with and without UBM particulate. RNA-Seq analysis was performed to identify changes in the transcriptome of these cells associated with exposure to UBM. While diabetic and non-diabetic cells exhibited different transcriptomes, these differences were minimized following incubation with UBM. ECs exposed to UBM exhibited changes in the expression of transcripts suggesting an increase in the endothelial–mesenchymal transition (EndoMT) associated with vessel maturation. Keratinocytes incubated with UBM demonstrated an increase in markers of activation. Comparison of the whole transcriptomes with public datasets suggested increased EndoMT and keratinocyte activation following UBM exposure. Both cell types exhibited loss of pro-inflammatory cytokines and adhesion molecules. These data suggest that application of UBM may accelerate healing by promoting a transition to the later stages of wound healing. This healing phenotype is achieved in cells isolated from both diabetic and non-diabetic donors.
1 INTRODUCTION
Proper wound healing is composed of four overlapping phases: haemostasis, inflammation, proliferation and remodelling.1 In diabetic subjects, the cells involved in the latter three stages (macrophages, endothelial cells [ECs] and keratinocytes) exhibit altered phenotypes that impair wound healing progression. For example, a persistent inflammatory state characterized by an increase in the ratio of M1 (pro-inflammatory) to M2 (anti-inflammatory) macrophages is present in the wounds of diabetic subjects.2, 3 In addition, ECs in diabetics have impaired angiogenic responses, and both ECs and keratinocytes display reduced proliferation and migration.4-6 As a result, progression to the final remodelling phase of wound healing is impeded in diabetic subjects.
Early in wound healing, both ECs and keratinocytes play an active role in the inflammatory phase of wound healing, releasing numerous cytokines to promote immune cell recruitment. ECs are the primary cells promoting wound angiogenesis.7 Proper angiogenesis requires a timely, proper balance between multiple angiogenic factors. Late in the angiogenic process, maturation and stabilization of vessels requires the loss of surplus vessels through apoptosis and endothelial–mesenchymal cell transition (EndoMT).8, 9 In the later stage of wound healing, keratinocytes become activated, entering a proliferative and migratory state and producing paracrine and autocrine signals to promote wound remodelling. As the wound resolves, keratinocytes return to their differentiated state.10 Type 2 diabetes is associated with a prolonged inflammatory phase and a delay in keratinocyte activation and reduced maturation of blood vessels.11-14
Porcine urinary bladder matrix (UBM) is a biomaterial that has the potential to prevent these phenotypic changes in ECs and keratinocytes in diabetic subjects, thereby promoting a normal healing response.3, 15-20 We recently reported acute exposure to UBM is associated with restoration of normal function of macrophages in diabetic wounds.3 Whether UBM exposure promotes a healing phenotype in other cells involved in the diabetic wound healing process remains to be seen.
Here, we report that the transcriptome of keratinocytes and ECs isolated from diabetic donors exhibit a pointedly altered transcriptome compared to similar cells from non-diabetic donors. Furthermore, exposure of these cells to UBM results in a significant shift in the transcriptome for both diabetic and non-diabetic ECs. The changes associated with exposure to UBM suggest attenuation of inflammation and facilitation of keratinocyte activation and blood vessel maturation. Thus, these data suggest that exposure to UBM can promote a healing phenotype in ECs and keratinocytes consistent with the later stages of wound healing. Together these data provide mechanistic information underlying the efficacy of UBM under diabetic settings and aid in developing pharmacodynamic tools for tracking its action in the clinical setting.
2 MATERIALS AND METHODS
2.1 Tissue culture
Human dermal ECs and human epidermal keratinocytes isolated from diabetic and non-diabetic donors were obtained from Lonza, Inc., and cultured in endothelial growth medium and keratinocyte growth medium, respectively, according to the manufacturer's instructions. Three lots from different donors of each cell type were obtained. UBM particulate was provided by ACell, Inc. Cells were seeded into 10 cm2 plates and incubated in the growth medium supplemented with 200 μg/mL of UBM particulate or vehicle for 18 h.
2.2 RNA-sequencing
Total RNA was prepared using the Qiagen miRNeasy minikit and the rRNA was depleted using the RiboMinus™ Eukaryote Kit (Invitrogen). A library was prepared using the TruSeq RNA kit followed by 1 × 100 bp sequencing using an Illumina HiSeq 2000 instrument. In collaboration with the Tulane Next Generation Sequence Analysis Core, RNA-Seq reads were aligned to the human genome (hg19 assembly) using Novoalign (Novocraft) and splice junctions were identified using TopHat. Coverage files and FPKM values were generated using SAMMate. Principal components analysis (PCA) was performed and differentially expressed genes (DEGs) (defined as transcripts exhibiting an adjusted p < 0.05) were identified using the DESeq2 package in R.21 Gene enrichment analysis was performed using the PANTHER overrepresentation test.22 The cell-specific list of total transcripts identified in each dataset was used as the background for the analysis. Only the most specific gene ontologies were considered. Gene ontologies specific to unrelated organ systems were not included in the analysis.
2.3 Similarity scores for ECs and fibroblasts
In order to determine in an unbiased manner whether the expression profile of our ECs switches away from a normal EC profile and towards a mesenchymal cell profile, we developed a similarity score based on gene expression profiles generated using the CellMapper R package.23 Genes enriched in cells expressing CD31 or FSP1 with a false discovery rate <0.001 in the EH171 microarray dataset of Expression Hub were identified (Tables S1 and S2).24 The counts for each of these cell-specific genes in our datasets were scaled by dividing the count for a given transcript in a given sample by the mean of the counts for that transcript across all samples. A similarity score for each individual sample was calculated as the median of the scaled counts across all transcripts for that sample.
2.4 Bulk RNA-Seq deconvolution to identify keratinocyte phenotype
Because of the similar expression profile between different keratinocyte phenotypes, a more robust method was need to determine changes in keratinocyte phenotype following UBM exposure. We used the MuSiC2 R package to perform bulk RNA-Seq cell type deconvolution using a publicly available human neonatal foreskin epidermis single cell RNA-Seq dataset (GSE147482) as a reference.25, 26 Eleven different cell types were identified in the GSE147482 dataset using the Seurat R package (Figure S1).27 This included eight different keratinocyte groups (one representing basal, three representing mitotic, one representing spinous and three representing granular keratinocytes). The percentage of cell types in keratinocytes incubated in the presence and absence of UBM was calculated for all keratinocyte phenotypes. Those cell types which had less than 1% representation were removed and the analysis was re-performed with only the basal and a single mitotic cell type. One sample was removed from the analysis as an outlier based on the Grubbs' test.
2.5 Droplet digital PCR
Differential expression analysis was confirmed using droplet digital RT-PCR. Total RNA was prepared from cell lysates as described above and mRNA encoding in the genes in Table S1 was measured using QX200 Droplet Digital PCR (Bio-Rad) coupled with appropriate Quantitect primer assays (Qiagen, Inc.) and the QX200 EvaGreen ddPCR Supermix (Bio-Rad).
3 RESULTS
3.1 Principal components analysis
Our goal was to determine whether incubation with UBM promotes a healing phenotype in keratinocytes and ECs obtained from diabetic donors. Transcriptome-wide level PCA was performed to determine the mechanisms underlying the variability between the transcriptomes of the ECs and keratinocytes incubated with and without UBM. Samples were clustered together according to the combination of diabetic status and exposure to UBM (Figure 1). Principal component 1, which represents the highest variability (84.7% in ECs and 95.6% in keratinocytes), was associated with exposure to UBM. UBM exposure was also associated with tighter clustering along principal component 2 compared to control in ECs. This finding suggests that differences in the transcriptome associated with diabetes were minimized following exposure to UBM. A similar finding was not seen in the keratinocytes, but it should be noted that this component only represented 2.2% of the variance, compared with 9.2% for the ECs, and the diabetic and non-diabetic keratinocytes clustered tightly together in the absence of UBM.
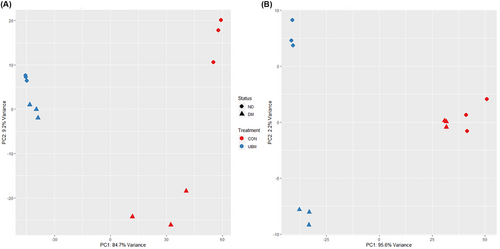
To better understand the mechanism underlying the variance in principal component 1, gene enrichment analysis was performed on the transcripts with loadings in the 5% most associated with UBM exposure, as seen as a negative shift in principal component 1. Table 1 lists the top biological processes exhibiting enrichment in the ECs and keratinocytes. Exposure to UBM was associated with enrichment in genes associated with the formation of desmosomes and hemidesmosomes. Additionally, genes directly associated with keratinization, cornification, and formation of the epidermis were enriched. While these findings were expected with keratinocytes, the findings of genes associated with cornification, establishment of skin barrier, and desmosome organization was unexpected in ECs. The inclusion of these gene ontologies was driven by lipases (cornification) and several genes associated with tight junctions (establishment of skin barrier and desmosome organization). The tight junction genes included non-canonical EC transcripts such as CLD1, CLD4 and DSP that are associated with de-differentiated ECs. These findings support that exposure to UBM promotes ECs and keratinocytes towards a phenotype that supports wound healing.
| Gene ontology | Fold enrichment | p-Value | False discovery rate | |
|---|---|---|---|---|
| Endothelial cell | Desmosome organization (GO:0002934) | 10.6 | 4.84E-06 | 4.90E-04 |
| Hyaluronan biosynthetic process (GO:0030213) | 10.09 | 2.42E-04 | 1.15E-02 | |
| Establishment of skin barrier (GO:0061436) | 9.32 | 1.10E-10 | 4.03E-08 | |
| Negative regulation of serine-type peptidase activity (GO:1902572) | 8.41 | 1.45E-03 | 4.65E-02 | |
| Hemidesmosome assembly (GO:0031581) | 7.85 | 7.42E-05 | 4.41E-03 | |
| Cornification (GO:0070268) | 7.61 | 3.63E-29 | 6.33E-26 | |
| Keratinization (GO:0031424) | 7.1 | 3.48E-34 | 1.09E-30 | |
| Keratinocyte | Desmosome organization (GO:0002934) | 10.39 | 5.63E-06 | 1.76E-03 |
| Establishment of skin barrier (GO:0061436) | 7.69 | 2.45E-08 | 2.02E-05 | |
| Negative regulation of epidermis development (GO:0045683) | 6.49 | 8.01E-05 | 1.51E-02 | |
| Cornification (GO:0070268) | 5.71 | 5.89E-19 | 1.15E-15 | |
| Phosphatidylethanolamine acyl-chain remodelling (GO:0036152) | 5.29 | 5.53E-05 | 1.11E-02 | |
| Regulation of keratinocyte proliferation (GO:0010837) | 5 | 1.89E-05 | 4.69E-03 | |
| Positive regulation of epidermis development (GO:0045684) | 4.81 | 6.36E-06 | 1.92E-03 |
3.2 Hierarchical clustering
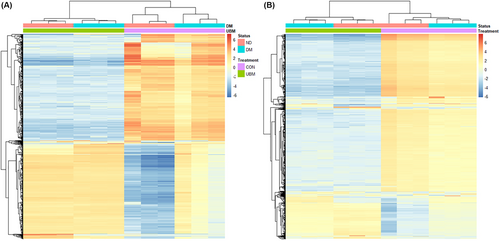
To better examine the similarities in the transcriptomes of the different samples, we performed hierarchical clustering of the 1000 most variable genes. In both the ECs and keratinocytes, the strongest grouping was according to exposure to UBM, with diabetic status being second (Figure 2). There was also a tighter clustering of the diabetic and non-diabetic samples exposed to UBM. The heatmaps for both cell types exhibited a large group of transcripts that were down-regulated with exposure to UBM compared to control, regardless of diabetic status. This suggests a common effect across the two phenotypes. The second-largest group of transcripts with common expression changes are those that are up-regulated with UBM exposure. Interestingly, while the response to UBM is similar, the control samples exhibited clear differences with the non-diabetic control samples exhibiting a reduced expression while the diabetic control samples exhibited a mild up-regulation. Overall, the hierarchical clustering suggests greater similarity between the transcriptomes of the UBM-exposed samples than is seen between the control samples.
3.3 Differential gene expression in ECs
A total of 14 760 DEG were identified between the ECs incubated with and without UBM with 6057 increased and 8703 decreased (Table S3). Diabetes is associated with stalled epithelial-mesenchymal transition (EMT) and poor angiogenesis. This association has been linked to extraordinarily high expression of the transcription factors ZEB1 and ZEB2.28 Genes encoding EC markers (PECAM1, VWF, ANGPT1, ANGPT2, FLT1 and TIE1) were uniformly down-regulated (Table 2). Similarly, the pro-angiogenic factors, VEGFA and TXN, were increased following exposure to UBM. Together, these suggest a role for EndoMT in the effects of UBM on diabetic ECs. The significantly down-regulated DEGs include several inflammatory mediators (PTX3, SRGN, TLR4, CCL2, CXCL2, ICAM1 and ICAM2). Additionally, an inhibitor endothelial nitric oxide synthase, RGS4, is dramatically reduced while XDH and NOS1 are increased. In contrast, NOS3 (eNOS) was actually reduced. Overall, this suggests a potential improvement in endothelial function.
| Gene ID | Log2 (fold change) | |
|---|---|---|
| Epithelial to mesenchymal transition | ZEB1 | −6.1 |
| ZEB2 | −7.1 | |
| LHX1 | 4.6 | |
| LHX2 | 4.8 | |
| PECAM1 | −11.4 | |
| VWF | −8.8 | |
| ANGPT1 | −3.6 | |
| ANGPT2 | −6.5 | |
| FLT1 | −7.9 | |
| TIE1 | −9.2 | |
| Angiogenesis | TXN | 1.3 |
| VEGFA | 2.3 | |
| Inflammation | CCL2 | −12.7 |
| CXCL2 | −2.2 | |
| ICAM1 | −7.7 | |
| ICAM2 | −9.0 | |
| IL6 | −6.2 | |
| TLR4 | −11.0 | |
| CCL14 | −9.5 | |
| CXCL8 | −7.3 | |
| CXCL1 | −1.8 | |
| CXCL10 | −5.8 | |
| CCR10 | −2.4 | |
| CXCR2 | 1.5 | |
| Endothelial dysfunction | RGS4 | −10.9 |
| XDH | 2.7 | |
| NOS1 | 2.2 | |
| NOS3 | −4.4 |
3.4 Differential gene expression in keratinocytes
A total of 15 662 DEG were identified between the keratinocytes incubated with and without UBM with 6536 increased and 9126 decreased (Table S4). Overall, the profile of the DEGs (Table 3) showed increases in genes associated with activation of keratinocytes (KRT6B, KRT6C, KRT16, TGFA and AREG) and a loss of genes associated with basal keratinocytes (TGFB2, ITGA5, ITGAV and ITGB1). The profile was not uniform, however, with KRT5, a marker of basal keratinocytes, being up-regulated and several genes associated with activated keratinocytes (CSF-2, CSF-3, FN1 and ICAM1) being down-regulated. Additionally, alterations in connexins associated with migratory keratinocyte phenotype were also seen as connexin-43 (GJA1) was decreased and connexins 26 (GJB2) and 30 (GJB6) were down-regulated. In addition to keratinocyte activation, a similar decrease in the expression of genes associated with inflammation (TLR4, CCL2, CXCL2, ICAM1, ICAM2, IL6, CCL14, CXCL8, CXCL10 and CXCR2) was observed following incubation with UBM. Together, this suggests UBM exposure was associated with a transition from an inflammatory state towards the activated state associated with wound remodelling.
| Gene ID | Log2 (fold change) | |
|---|---|---|
| Inflammation | TLR4 | −12.5 |
| CCL2 | −9.4 | |
| CXCL2 | −1.0 | |
| ICAM1 | −4.2 | |
| ICAM2 | −7.2 | |
| IL6 | −3.7 | |
| CCL14 | −8.9 | |
| CXCL8 | −4.0 | |
| CXCL10 | −3.8 | |
| CXCR2 | 1.2 | |
| Activated keratinocytes | KRT6A | 1.0 |
| KRT6B | 1.1 | |
| KRT6C | 1.4 | |
| KRT16 | 1.4 | |
| KRT17 | 1.0 | |
| WEE1 | 1.4 | |
| TGFA | 1.6 | |
| AREG | 1.6 | |
| CSF2 | −4.1 | |
| CSF3 | −5.0 | |
| FN1 | −1.8 | |
| Basal keratinocytes | TGFB2 | −2.5 |
| ITGA5 | −1.6 | |
| ITGAV | −1.8 | |
| KRT5 | 1.1 | |
| Gap junction regulation | GJA1 | −0.8 |
| GJB2 | 1.4 | |
| GJB6 | 2.7 |
3.5 EndoMT in ECs following UBM exposure
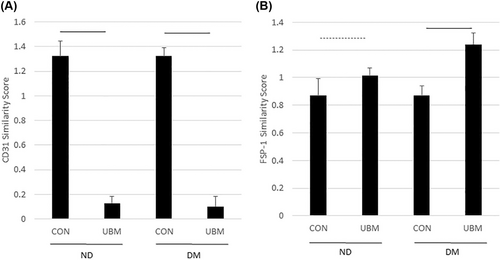
Analysis of the DEGs from the ECs suggested that EndoMT may occur following UBM exposure. To further examine this possibility, we developed a score to measure the similarity of our dataset to publicly available data from cells expressing markers of ECs (CD31) and fibroblasts (FSP1) and compared this score across the EC groups. As shown in Figure 3A, the score for similarity to CD31 expressing cells significantly decreases following UBM exposure in the ECs isolated from both diabetic and non-diabetic donors. The score for similarity to cells expressing FSP1 was increased in both sets of ECs following UBM exposure with the diabetic group exhibiting a significant increase (Figure 3B). These findings further support the possibility that ECs undergo EndoMT following UBM exposure.
3.6 Increase in mitotic keratinocytes following UBM exposure
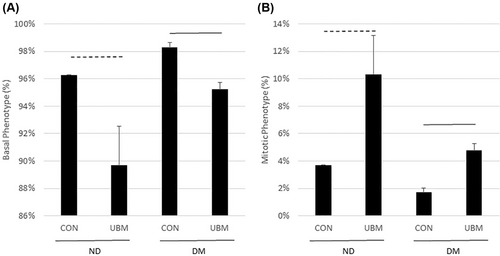
The DEGs from the keratinocyte dataset suggested there is an increase in keratinocyte proliferation following UBM exposure. To determine whether there was a change in keratinocyte phenotype following UBM exposure, the percentage of different keratinocyte phenotypes was determined using bulk RNA-Seq deconvolution. As seen in Figure 4, in both the diabetic and non-diabetic groups the percentage of basal keratinocytes decreases and the mitotic keratinocytes increase, suggesting increased proliferation.
4 DISCUSSION
Multiple case studies have established that treatment with UBM accelerates healing in a variety of wound types, including severe chronic wounds, traumatic wounds and radiation wounds.29-32 Such reports demonstrate the breadth of wound types for which UBM represents an effective treatment option. These studies have not, however, specifically addressed diabetes as a co-morbidity. We have previously reported that UBM promotes restoration of a normal ratio of macrophage phenotypes in diabetic wounds.3 It is well established that diabetes hinders wound healing, in part through decreased keratinocyte activation and impaired maturation of blood vessels.11-14 This study was undertaken to assess whether, similar to macrophages, exposure of ECs and keratinocytes of diabetic donors to UBM induces phenotypic changes more conducive to wound healing.
Comparison of the full transcriptomes of the diabetic and non-diabetic keratinocytes and ECs using PCA and hierarchical clustering revealed a clear response to exposure with UBM. PCA analysis demonstrated that both ECs and keratinocytes clustered according to the diabetic status of the donor. It also demonstrated that UBM exposure produces a far greater difference in transcript expression than the difference in phenotype. As UBM has been shown to accelerate healing,3, 29-32 this difference likely represents the transition to a healing phenotype by both the diabetic and non-diabetic cells. Hierarchical clustering of the top 1000 most variable genes indicates that exposure to UBM is associated with a down-regulation of a large number of genes in both diabetic and non-diabetic phenotypes. The second-largest group of transcripts in the clustering is that which is up-regulated in response to UBM exposure in both cell types. Interestingly, these transcripts exhibit lower expression in the non-diabetic cells, but slightly elevated expression in the diabetic subjects. This finding demonstrates that despite a clear difference in expression under control conditions, UBM exposure leads to a similar transcriptional profile.
As part of proper wound healing, keratinocytes are activated and enter a proliferative state. Additionally, the keratinocytes begin to differentiate towards the formation of the cornified envelope.33 Acellular dermal matrices have been shown in animal models to promote increased proliferation of keratinocytes.34 Our in vitro data demonstrate that human keratinocytes express multiple markers of activation and exhibit a transcriptome with greater mitotic character following acute exposure to UBM. Together, these data suggest UBM exposure promotes keratinocyte activation and, in turn, development of the cornified envelope. It also suggested potential changes in reactive oxygen species production through decreased NOX4 expression.
ECs are critical to the angiogenesis that occurs in wound healing. Early in wound healing, an excess of neovascularization occurs. Late in the process, the unneeded vessels disappear. This process is believed to occur largely through apoptosis. Recently, EndoMT has also been proposed to play a role in this maturation of the blood vessels. This transition is impaired in the wounds of diabetic patients. Both the DEGs and our similarity scores suggest that ECs exhibit an increase in EndoMT. An increase in EndoMT was also observed in wounds treated with poly (caprolactone) gelatin nanofibrous composite scaffold containing silicate-based bioceramic particles, a scaffold engineered to accelerate diabetic wound healing.35
Both cell types demonstrated a loss of pro-inflammatory transcripts following exposure to UBM. UBM and other bioscaffolds have been found to promote decreased inflammation.17-19, 36, 37 Impaired wound healing in type 2 diabetes is associated with prolonged inflammation. Previously, we have reported that exposure to UBM promoted a reduction in the pro-inflammatory nature of the diabetic wound.3 These data suggest a similar response to UBM in other cell types that play an important role in the wound healing process.
In this report, we present transcriptome comparisons of diabetic and non-diabetic ECs and keratinocytes that suggest that diabetes does induce a unique phenotype and that exposure to UBM yields a dramatic change in phenotype. As UBM is associated with accelerated wound healing, we suggest that this represents the transition towards a healing phenotype. Further, the data suggest a common response in both diabetic and non-diabetic cells, implying that UBM promotes this healing phenotype regardless of the diabetic status. As an in vitro study, additional studies are needed to confirm whether these findings occur clinically. Additionally, this study was limited to RNA data, so confirmation of these effects at the protein or cellular level is needed. Nonetheless, this study provides a basis for a better understanding of UBM ability to promote wound healing in both diabetic and non-diabetic patients through influences on ECs and keratinocytes.
AUTHOR CONTRIBUTIONS
John T. Paige and T. Cooper Woods designed the experiments. Daniel J. Lightell, Hunter F. Douglas, Natasha Klingenberg and Thaidan Pham performed the experiments and data analysis. T. Cooper Woods analysed the RNA-Seq data. John T. Paige and T. Cooper Woods drafted the manuscript.
ACKNOWLEDGEMENTS
This research was supported through an investigator-initiated grant from ACell, Inc. and by the National Institute of General Medical Sciences and the National Heart Lung Blood Institute of the National Institutes of Health under Award Numbers R01HL127092 and P30GM103337.
CONFLICT OF INTEREST STATEMENT
This research was supported through an investigator-initiated grant from ACell, Inc., the manufacturer of the UBM.




