R164C mutation in FOXQ1 H3 domain affects formation of the hair medulla
Abstract
A number of single gene mutations in laboratory mice produce hair follicle defects resulting in deformed hair shafts. The radiation-induced (SB/LeJ-Foxq1sa) satin mutant mice have a satin-like sheen to their hair and dilute colouration. This sheen is due to failure of the hair shafts to develop normal medullas, while the pigment dilution is due to the unrelated beige (lysosomal trafficking regulator, Lystbg) mutation. A new allelic mutation, Foxq1sa-J, arose spontaneously on the albino (tyrosinase, Tyrc) MRL/MpJ-Faslpr background. The Foxq1sa-J allele has a C to T transition at position 490. By contrast, the Foxq1sa mutant allele was confirmed to be a 67 base pair deletion followed by two base changes (GA to AT). Morphologic changes were similar to those seen in Hoxc13 transgenic and targeted mutant mice. This new allelic mutation provides yet another tool to investigate formation of the interior structures of hair shafts.
Abbreviations
-
- Foxq1
-
- forkhead box Q1 gene
-
- Foxq1sa
-
- satin allele
-
- Foxq1sa-J
-
- satin-J allele
-
- Foxq1sa-e1
-
- satin-e1 allelic mutation
-
- SEM
-
- scanning electron microscopy
-
- TEM
-
- transmission electron microscopy
Background
The satin mutation is due to a deletion in the Foxq1 gene 1. A second allele, Foxq1sa-e1, arose in an ENU mutagenesis screen 1. A third mutation, satin-J (Foxq1sa-J), arose spontaneously in the albino MRL/MpJ-Faslpr colony. All three fail to develop a hair shaft medulla. The Foxq1sa-J allele was found to be allelic with Foxq1sa 2. We report here the phenotype and molecular defect of this new allelic mutation.
Questions Addressed
This study aimed to identify the molecular defect and contrast the phenotype of a new, spontaneous allelic mutation of the Foxq1 gene (Foxq1sa-J) to the commonly used satin allele.
Experimental Design
All studies were approved by The Jackson Laboratory Animal Care and Use Committee (ACUC). To define histological lesions in the hair, 5-day-old mice were used when hair follicles were in late anagen. Scanning electron microscopy (SEM), light and polarized light microscopy were performed on pelage hair samples from two each of SB/LeJ-Foxq1sa/Foxq1sa, MRL/MpJ-Faslpr and MRL/MpJ-Faslpr-Foxq1sa-J/Foxq1sa-J mice to compare medullary defects 3, 4.
Mutations in the Foxq1 gene were identified by sequence comparison between MRL/MpJ-Faslpr-Foxq1sa-J/Foxq1sa-J mutant mice and wild-type control mice.
Results
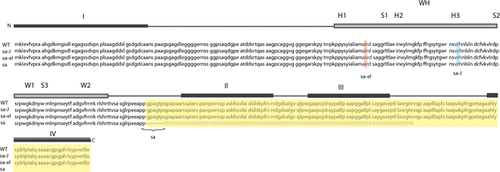
In the Foxq1sa-J ORF, there was a novel C to T transition at position 490 (Figure S1). This should result in an arginine (R) to cysteine (C) conversion at amino acid (aa) 164 (R164C). The previously reported 67 base deletion and downstream successive base changes(GA to AT) at positions 766 and 767 in the Foxq1sa allele were also confirmed 1. The deletion causes a truncation of the protein after aa 128 affecting the transcriptional activity of the protein, but not its DNA-binding capacity in contrast to the Foxq1sa-J allele.
The DNA-binding domain of FOXQ1 consists of two wings (W1 and W2), three α-helices (H1, H2 and H3) and three β-strands (S1, S2 and S3), arranged in the following order, H1-S1-H2-H3-S2-W1-S3-W2 (Fig. 1) 5, 6. The Foxq1sa-e1 and Foxq1sa-J mutations affect the C-terminal H1 and H3 domains, respectively (Fig. 1). The resulting amino acid changes result in different protein polarity that is predicted to destabilize DNA-protein interactions (Fig. 1). By contrast, the Foxq1sa mutation occurs downstream from the DNA-binding domain, which is predicted to interfere with transcriptional activation and/or repression activities 1, 7 (Fig. 1). Thus, three mutations affecting different parts of the FOXQ1 protein lead to very similar phenotypes.
The Foxq1sa and Foxq1sa-J mutant mice have a satiny hair sheen due to refraction of light through the defective hair shafts (Fig. 2a-c). No structural abnormalities were noted on the external surface of the mutant hair shafts by scanning electron microscopy (Fig. 2d-f). Light microscopy reveals normal medullary septation and septulation patterns in wildtype mice (Fig. 2h,k), but loss of these patterns in Foxq1sa and Foxq1sa-J mutant mice (Fig. 2g,i,j,l). Differences between the hairs of these two allelic mutations are limited to pigment clumping resulting from the Lystbg mutation in Foxq1sa mice (Fig. 2g,j,m;arrowheads) and the Tyrc mutation in the Foxq1sa-J mouse.
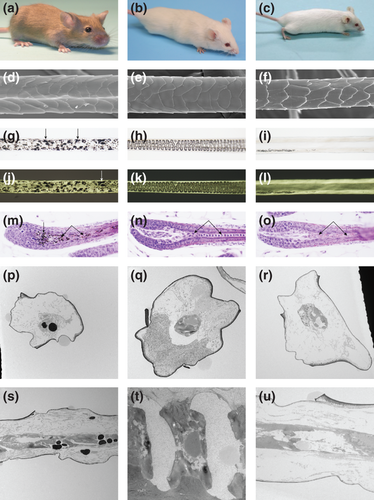
Histological examination of hair follicles revealed that the normally well-organized, compressed cells of the premedulla and medulla (Fig. 2n) formed in a disorganized manner in satin mutant mice. Rather than the nuclei being flattened parallel to the epidermis, the mutant hair medulla nuclei were elongated perpendicular to the epidermis (Fig. 2m,o). By transmission electron microscopy, normal septation and septulation patterns of hair shafts from the control mice (Fig. 2q,t) were contrasted with the loss of these structures in the two satin allelic mutations (Fig. 2p,s,r,u). Wild-type and heterozygous Foxq1sa-J hairs were clinically normal with well-formed hair medullas. Longitudinal sections revealed regularly patterned, rectangular cells in the medulla with clear spaces representing contraction artifact (data not shown). Both the Foxq1sa (Fig. 2p,s) and Foxq1sa-J (Fig. 2r,u) hairs had cells in the medulla that were poorly preserved. In control mice, these cells were formed regular, rectangular structures. By contrast, both satin mutant mice had medulla cells that lost this regular rectangular medullary cell pattern; instead, the cells were elongated and filled the cavity (Fig. 2p,s).
Conclusion
A vast array of signalling pathways operate in the developing hair follicle, whose coordination is essential for proper hair formation 8-10. Yet, the molecular mechanisms governing these processes are still largely unknown. We report here a new publicly available, spontaneous mutant (Foxq1sa-J) that results in similar hair sheen and medullation defects as the Foxq1sa allele 1.
Foxq1 is expressed in the upper bulb of the anagen hair follicle 11, a region in which keratinocytes undergo patterns of differentiation to form the respective elements of the hair shaft and follicle 12. Foxq1 is one of many genes whose mutations in mice are linked to hair shaft defects including Foxn1nu 13, Hoxc13 14 and Dsg4 15. Both Foxq1 and Foxn1 are transcription factors and targets of HOXC13 regulation 11, 16. In light of these mutations, we hypothesize that these genes work in a coordinate manner to direct development and cycling of the hair shaft.
Acknowledgements
The authors thank J. Hammer for assistance with the graphics and L.S. Bechtold with electron microscopy.
Author contributions
Author contributions are as follows: B.W., K.A.S, V.K., J.P.S. performed the research, B.W., J.P.S. designed the study, B.W., C.H.P, C.S.P, J.P.S. analysed the data and wrote the paper.
Funding
This work was supported by the National Institutes of Health (AR053639, AR056635, RR00173, and CA34196), National Natural Scientific Foundation of China (31071092) and mentorship grants from the North American Hair Research Society. Shared scientific services were supported in part by a Basic Cancer Center Core Grant from the National Cancer Institute (CA34196).
Conflict of interests
The authors have declared no conflicting interests.




