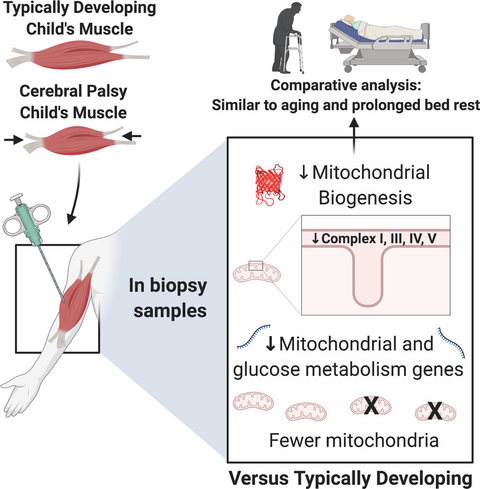
Reduced mitochondrial DNA and OXPHOS protein content in skeletal muscle of children with cerebral palsy
Abstract
Aim
To provide a detailed gene and protein expression analysis related to mitochondrial biogenesis and assess mitochondrial content in skeletal muscle of children with cerebral palsy (CP).
Method
Biceps brachii muscle samples were collected from 19 children with CP (mean [SD] age 15y 4mo [2y 6mo], range 9–18y, 16 males, three females) and 10 typically developing comparison children (mean [SD] age 15y [4y], range 7–21y, eight males, two females). Gene expression (quantitative reverse transcription polymerase chain reaction [PCR]), mitochondrial DNA (mtDNA) to genomic DNA ratio (quantitative PCR), and protein abundance (western blotting) were analyzed. Microarray data sets (CP/aging/bed rest) were analyzed with a focused query investigating metabolism- and mitochondria-related gene networks.
Results
The mtDNA to genomic DNA ratio was lower in the children with CP compared to the typically developing group (−23%, p=0.002). Out of five investigated complexes in the mitochondrial respiratory chain, we observed lower protein levels of all complexes (I, III, IV, V, −20% to −37%; p<0.05) except complex II. Total peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α) messenger RNA (p<0.004), isoforms PGC1α1 (p=0.05), and PGC1α4 (p<0.001) were reduced in CP. Transcriptional similarities were observed between CP, aging, and 90 days’ bed rest.
Interpretation
Mitochondrial biogenesis, mtDNA, and oxidative phosphorylation protein content are reduced in CP muscle compared with typically developing muscle. Transcriptional pathways shared between aging and long-term unloading suggests metabolic dysregulation in CP, which may guide therapeutic strategies for combatting CP muscle pathology.
What this paper adds
- Cerebral palsy (CP) muscle contains fewer energy-generating organelles than typically developing muscle.
- Gene expression in CP muscle is similar to aging and long-term bed rest.
What this paper adds
- Cerebral palsy (CP) muscle contains fewer energy-generating organelles than typically developing muscle.
- Gene expression in CP muscle is similar to aging and long-term bed rest.
We compared skeletal muscle samples from children with cerebral palsy (CP) and typically developing children and observed evidence of reduced mtDNA and OXPHOS protein content in CP skeletal muscle, indicating reduced mitochondrial abundance. Moreover, transcriptional similarities to aging and long-term unloading suggests signs of metabolic alterations in CP skeletal muscle.
Abbreviations
-
- mtDNA
-
- Mitochondrial DNA
-
- OXPHOS
-
- Oxidative phosphorylation
-
- PCR
-
- Polymerase chain reaction
-
- PGC
-
- Peroxisome proliferator-activated receptor gamma coactivator
-
- Tfam
-
- Transcription factor A
Cerebral palsy (CP) is the most common cause of motor impairment in children.1 Although the initial brain injury is non-progressive, skeletal muscle function typically deteriorates over time.2 Skeletal muscles of individuals with CP are small, weak, and compositionally different compared with typically developing muscle. Specifically, increased collagen content3 and intramuscular adipose tissue4 are believed to contribute to poor muscle function in this population. Poor muscle quality is a well-described risk factor for insulin resistance and development of type 2 diabetes5, 6 and is typically associated with a reduced mitochondrial content of the muscle.7 Whether the described pathological findings in CP muscle influences metabolic capacity of the tissue is not known.
A recent study by Zogby et al. investigated skeletal muscle mitochondrial characteristics in CP muscle using an immunohistochemical approach. According to fiber-type-specific succinate dehydrogenase activity,8 their results showed a similar succinate dehydrogenase activity per unit area in both type I and type II fibers in individuals with CP and typically developing individuals, suggesting that muscle oxidative potential is preserved in children with CP. Conversely, other studies reporting on fiber types in the muscles of children with CP describe a clear slow to fast-fiber-type shift,9, 10 indicative of a less oxidative phenotype of the muscle, as fast type IIx fibers contain fewer mitochondria than slow type I fibers. Moreover, two skeletal muscle transcriptomics studies in children with CP11, 12 indicate reduced messenger RNA levels of key genes involved in skeletal muscle mitochondrial biogenesis, such as AMP-activated protein kinase (AMPK) and transcription factor A (Tfam), contrasting with the data of Zogby et al. An accurate picture of the metabolic characteristics of CP muscle is of great importance for developing the appropriate therapeutics to counteract muscle dysfunction.
Given the discrepancies in the literature, we wanted to use genomic, transcriptomic, targeted messenger RNA, and protein-based approaches to investigate whether: (1) mitochondrial DNA (mtDNA) abundance and protein levels of oxidative phosphorylation (OXPHOS) enzymes, as markers of mitochondrial content, are reduced in CP muscle compared with typically developing muscle; (2) gene and protein expression of factors involved in mitochondrial biogenesis are reduced in CP muscle compared with typically developing muscle; and (3) gene networks linked to mitochondrial biology and glucose metabolism are downregulated in CP muscle compared with typically developing muscle. In addition, CP gene networks were compared with severe inactivity and aging data sets to identify common themes with respect to metabolic dysregulation and provide translatable insight into appropriate therapeutic targets.
METHOD
Participants
Nineteen children and adolescents (mean [SD] age 15y 4mo [2y 6mo]; range 9–18y; 16 males, three females) with CP, scheduled for surgical lengthening of the biceps brachii tendon, were included in the study. All but one patient had a developed contracture of the elbow flexor muscles, resulting in an extension deficit of the elbow. Their gross motor function was classified on a five-level scale according to the Gross Motor Function Classification System, and their ability to use their hands was classified on a five-level scale according to the Manual Ability Classification System. For details on type of CP, clinical presentation (unilateral/bilateral), sex, age, contracture, Gross Motor Function Classification System level, and Manual Ability Classification System level, see Table S1 (online supporting information). Skeletal muscle samples were obtained intraoperatively under general anesthesia and were frozen in isopentane cooled on liquid nitrogen. All children had fasted for a minimum of 10 hours before surgery. Control biceps brachii muscle samples were obtained post-mortem from children and young adults who had sustained accidental deaths (n=10, mean age 15y 1mo; range, 7–21y; two females and eight males; Table S2, online supporting information). All samples were stored in a freezer at −80°C until analysis. Seventeen out of 19 samples were available for DNA analysis and 14 out of 19 samples for gene expression, although not all samples were from the same individuals. Owing to limited muscle material, 9 out of 19 samples and 11 out of 19 samples were available for OXPHOS and Tfam protein analysis respectively.
RNA/DNA extraction, cDNA synthesis, and quantitative polymerase chain reaction
Skeletal muscle biopsies (˜25mg) were homogenized in TRIzol reagent (Invitrogen, Carlsbad, CA, USA) using a handheld homogenizer (Omni International, Kennesaw, GA, USA). RNA and DNA were extracted according to the information provided by the manufacturer (Invitrogen). RNA was quantified using a NanodropVR 2000 (Thermo Scientific, Gothenburg, Sweden), and integrity assessed by agarose gel electrophoresis. DNA was resuspended in 8mM NaOH, then centrifuged at 4°C at 12 000g to remove insoluble material. The DNA was transferred to a new tube and pH adjusted with HEPES. All samples were stored at −20°C until further use. Five hundred nanograms of RNA were reverse-transcribed with a VILO cDNA synthesis kit (Invitrogen) according to the manufacturer’s recommendations. Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) was used for quantitative polymerase chain reaction (PCR) in a QuantStudio 3 Real-Time quantitative PCR Systems machine (Thermo Fisher Scientific, Waltham, MA, USA). Quantitative PCR data (total peroxisome proliferator-activated receptor gamma coactivator 1-alpha [PGC1α] and splice variants PGC1-α1, α2, α3, and α4)13 were normalized to the geometric mean of three stable reference genes (GAPDH, EMC7, VCP). Primer sequences are available in Table S3 (online supporting information). Melting curves were performed for every primer pair to confirm a single-product amplification. All samples were run in duplicates and quantitative reverse transcription PCR data were analyzed using the 2−ΔΔCT method.
Quantification of mtDNA to genomic DNA
DNA was diluted to a concentration of 10ng/μL. Primers for the myogenin promoter was used to quantify genomic DNA and the mitochondrial gene cytochrome c oxidase I for mtDNA as previously described.14 Quantitative reverse transcription PCR was performed and analyzed as described for muscle gene expression. Primer sequences are available in Table S3.
Western blotting
Muscle samples (approximately 20mg) were homogenized in radio-immuno-precipitation assay buffer (20μl radio-immuno-precipitation assay/mg tissue) containing 150mM NaCl; 10mM Tris-HCl (pH 7.8); 5mM ethylenediaminetetraacetic acid (pH 7.4); 0.5% Na deoxycholate; 0.1% sodium dodecyl sulfate; 1% Triton-X100; and 1× complete protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland) using glass homogenizers. The homogenate was rotated gently at 4°C for 60 minutes, followed by centrifugation at 4°C for 10 minutes (15 000g). All chemicals were purchased from Sigma unless stated otherwise (Sigma-Aldrich, St Louis, MO, USA). Protein concentration was assessed with the detergent compatible protein assay (Bio-Rad, Hercules, CA, USA) and lysates diluted (if needed) with lysis buffer before mixing 3:1 with 4× Laemmli buffer (Bio-Rad) containing 10% β-mercaptoethanol. All samples were heat-denatured for 10 minutes, cooled on ice, and stored at −20°C until further use. Skeletal muscle homogenates (25μg protein/sample) were separated by electrophoresis on 4% to 15% TGX criterion sodium dodecyl sulfate–PAGE gels (Bio-Rad) and proteins were then transferred onto polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked at room temperature with Everyblot blocking buffer (Bio-Rad) and then incubated with primary antibodies for Tfam (1:1000, no. 850501) from BioLegend (San Diego, CA, USA). The OXPHOS cocktail (1:1000, no. 110411) from Abcam (Cambridge, UK) was used to blot for OXPHOS proteins. All primary antibodies were diluted in Everyblot blocking buffer and incubated overnight at 4°C. After washing with tris buffered saline with Tween 20 (0.1%), membranes were incubated for 1 hour at room temperature with goat anti-mouse secondary antibody (1:10 000 in blocking solution, no. 31430) from Invitrogen. After washing steps with tris buffered saline with Tween 20, membranes were incubated for 5 minutes with enhanced chemiluminescence reagent (Clarity Western ECL substrate, #170-5060, Bio-Rad) and exposed using a ChemiDoc MP Imaging System (Bio-Rad). Immunoblots were quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA).15 Coomassie blue staining was used to confirm equal loading and for densitometry normalization.
Secondary analysis of publicly available data sets
Publicly available skeletal muscle microarray data sets for CP (GSE31243),12 aging (GSE38718),16 and bed rest (GSE148152)17 were used to determine shared signatures in gene expression. The sampled muscles were not the same in these studies: CP muscle (hamstrings muscle), aging (biceps brachii), and bed rest (vastus lateralis muscles). Raw data were downloaded and reanalyzed using packages from the Bioconductor consortium. For each array, raw intensity was preprocessed and normalized with robust microarray analysis. Differentially expressed genes were detected using a linear model implemented in R package limma version 3.32.7 (Linear Model for Microarray Data; R Foundation for Statistical Computing, Vienna, Austria).18 The p-values were adjusted for multiple testing by the Benjamini–Hochberg method to control the false discovery rate. Differentially expressed genes were considered when the adjusted p-value was ≤0.05. Differentially expressed gene lists were uploaded to g:Profiler for pathway analysis and uploaded to Cytoscape version 3.8 for subsequent enrichment analysis and data visualization.19 Nodes were generated with an adjusted p<0.05 for enrichment. The size of each node was scaled to the gene set size; the shape of each node is specific to gene ontologies (circle) or pathway (diamond) enrichment. The color of the node was scaled to the adjusted p-value. Edges connecting nodes were scaled to the similarity coefficient (genes shared between nodes).
Statistical analysis
Statistical analysis was performed with Prism 8 (GraphPad Software, CA, USA). Values are reported as mean (SD). Differences in messenger RNA transcript levels, mtDNA to genomic DNA ratio, and protein levels were assessed by a Student’s t-test (data normality investigated with a Shapiro–Wilk test). Significance level was set at p<0.05 for a two-sided t-test.
Ethics
The ethical review board of Karolinska Institutet, Stockholm, Sweden, approved the study (Dnr 01-012, addendum 2018/1739-32, 04-324/2). The autopsy specimens were collected in agreement with Swedish laws and regulations on autopsy and transplantation with approval by the National Board of Health and Welfare. Informed written consent was obtained from all participants with CP and the parents of those younger than 18 years.
RESULTS
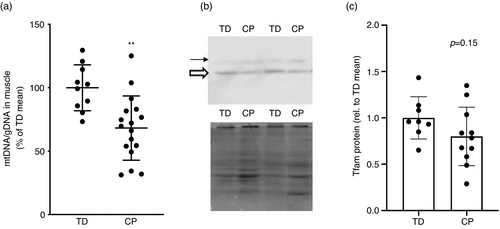
To investigate oxidative capacity in CP muscle, we quantified levels of five OXPHOS proteins: complex I (NDUFB8), complex II (SDHB), complex III (UQCRC2), complex IV (COXII), and complex V (ATP5A). Four out of five complexes (I, III, IV, V) showed a significant reduction of −20% to −37% (p<0.05) in content in CP muscle, whereas complex II tended to be lower in CP but was not statistically different to typically developing muscle (−33%, p=0.07; Fig. 1). We observed a lower mtDNA to genomic DNA ratio (−23%, p=0.002) in CP biceps brachii muscle compared with typically developing biceps brachii muscle (Fig. 2a). Skeletal muscle protein levels of Tfam were not significantly different in CP muscle compared with typically developing muscle (p=0.15, Fig 2b,c).

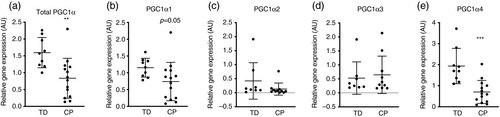
Consistent with evidence of reduced mitochondrial content, PGC1α gene expression was suppressed (−24%, 1.60 [0.45] vs 0.84 [0.55], p=0.004) in CP muscle compared with typically developing muscle (Fig. 3a). Splice variants displayed a non-homogenous expression in CP muscle; PGC1α1 and PGC1α4 were reduced (−36%, 1.15 [0.28] vs 0.74 [0.54], p=0.05; −64%, 1.94 [0.84] vs 0.70 [0.53], p<0.001 respectively) compared with typically developing muscle, whereas PGC1α2 (−69%, 0.42 [0.65] vs 0.13 [0.29], p=0.18) and PGC1α3 (+22, 0.53 [0.58] vs 0.65 [0.82], p=0.70) were not (Fig. 3b–e).
A secondary analysis of a publicly available microarray data set comparing CP and typically developing hamstrings muscles12 produced 2290 significantly downregulated genes in CP muscle. Gene ontology and pathway analyses revealed glycolytic and mitochondrial transcriptional networks to be suppressed in CP skeletal muscle (Fig. 4a,b). The gene set enriching the mitochondrial biogenesis node is shown in Figure 4c and the gene set enriching the glycolytic transcriptional network is shown in Figure 4d. We also compared the CP skeletal muscle transcriptome with that of older adults16 and individuals subjected to prolonged bed rest17 to determine whether a similar metabolic signature existed between conditions. CP shared more genes with the aging data set (332 genes) compared with the bed rest data set (109 genes). We found that 28 downregulated genes were shared between the three conditions (Fig. S1, online supporting information) and that mitochondrial and aerobic respiration transcriptional networks were suppressed under all three conditions (Fig. S2a,b, online supporting information), including gene ontologies (Reactome) such as mitochondrial translation initiation, elongation, and termination (MRPS15, MRPL2, MRPL35) and tricarboxylic acid cycle and electron transport chain (CYCS, OGDH, SLC16A1, ATP5MPL, NNT, DLAT). For a complete list of genes shared between CP and/or aging/bed rest see Table S4 (online supporting information).

DISCUSSION
This study addresses biological processes involved in mitochondrial biogenesis, mtDNA content, and OXPHOS protein abundance in CP skeletal muscle. We show that key upstream regulators of metabolism and mitochondrial biogenesis are reduced in CP skeletal muscle compared with typically developing controls, and that this is associated with downregulation of large gene networks affecting glucose metabolism and mitochondrial biology. Skeletal muscle in individuals with CP also contains lower amounts of mtDNA, potentially indicating fewer mitochondria in CP skeletal muscle compared with typically developing muscle. Moreover, we also observed lower levels of OXPHOS proteins in CP skeletal muscle that, together with reduced overall mitochondrial content, suggests a reduced oxidative capacity in CP muscle compared with typically developing muscle.
PGC1α is a key regulator of mitochondrial biogenesis in skeletal muscle.20 Transgenic mice that overexpress PGC1α in skeletal muscle display an oxidative phenotype with higher exercise tolerance than control mice and greater mitochondrial content.21, 22 On the contrary, several studies that have used transgenic mice lacking PGC1α have reported intact exercise adaptations including mitochondrial biogenesis,23-25 indicating a high level of biological complexity that is still not fully elucidated. Acute exercise studies in humans have also identified PGC1α as an early response gene to aerobic exercise, preceding mitochondria-related gene expression.26 PGC1α indirectly influences mtDNA transcription via increased expression of mitochondrial Tfam, which is coactivated by nuclear respiratory factor-1.20 In our study, we observed reduced abundance of both total levels of PGC1α and isoforms PGC1α1 and PGC1α4. PGC1α1 is believed to be the splice isoform most involved in mitochondrial biogenesis,27 whereas PGC1α4 has been implicated in skeletal muscle size regulation.13 Contrasting data from two previous skeletal muscle transcriptomics studies in children with CP,11, 12 we did not observe a significant reduction in Tfam protein in skeletal muscle potentially owing to low sample number for this specific analysis. In conjunction with these data, we observed lower levels of mtDNA/genomic DNA, indicating fewer mitochondria in CP skeletal muscle than in typically developing muscle. Our skeletal muscle data are supported by recent findings that children with CP also have reduced mitochondrial content in peripheral blood cells.28 Blood-based bioenergetic profiling correlates to physical function in older individuals29 and has been suggested as a proxy for skeletal muscle oxidative capacity.30 Taken together, our data suggest a reduced activity of mitochondrial biogenesis in individuals with CP compared with typically developing children.
Mitochondrial content in skeletal muscle has been linked to aerobic capacity31, 32 and shown to increase in response to aerobic training.33 Individuals with CP have lower VO2 max than typically developing peers.34, 35 Aerobic capacity depends on central (cardiac output, ventilation) and peripheral (perfusion, oxygen extraction, mitochondrial content) factors. For individuals with CP, it is mostly unknown how these parameters are influenced by increased exercise intensity and thereby an increased demand for energy (i.e. ATP). Interestingly, lactate production is higher than expected at moderate exercise intensities in individuals with CP, indicating that an insufficient ATP production is evident already at relatively light exercise intensities.36 The knowledge about metabolic capacity of CP skeletal muscle is scarce. Transcriptomics studies have indicated reduced expression of gene ontologies related to oxidative metabolism.11, 12 We observed reduced levels of four out of five electron transport chain complexes (I, III, IV, V) with only succinate dehydrogenase (II) not reaching statistically significant levels. Interestingly, complex II does not include any mtDNA encoded subunits, tentatively suggesting that reduced mtDNA content primarily affects the other complexes more as they are dependent on mitochondrially translated proteins. Our data support the previous observation of a reduced aerobic capacity in skeletal muscle and matches previously published fiber-type data from CP biceps muscle that shows a shift to faster myosin isoforms (IIx).10 Thus, our data suggest a role for decreased skeletal muscle mitochondrial content and potentially also capacity in the reduced exercise tolerance typically observed in individuals with CP.
Early metabolic dysregulation of skeletal muscle most likely plays a role in the increased risk of metabolic and cardiovascular disease that has been observed in the adult population with CP.37, 38 To gain further insight into metabolism-related gene expression in CP skeletal muscle, we performed a secondary analysis of publicly available hamstrings muscle microarray data. A gene ontology analysis (biological processes, reactome) on all significantly downregulated genes showed that glucose metabolism- and mitochondria-related gene networks were enriched in CP muscle. Surprisingly, CP muscle shared more downregulated genes with aged skeletal muscle than with long-term disuse. This is intriguing and challenges the belief that CP skeletal muscle pathophysiology is due to disuse per se and suggests common mechanisms with aging, most likely involving altered skeletal muscle neural input. We found that 28 downregulated genes were shared between the three conditions. Mitochondrial and aerobic respiration transcriptional networks were suppressed under all three conditions and out of the 28 common genes, 14 had mitochondrial localization as indicated by Mitocarta 3.0.39 This focused analysis on gene expression supports the biochemical data presented in this paper and further suggests that individuals with CP, already at a young age, display evidence of metabolic dysregulation in skeletal muscle, as previously described with aging and long-term bed rest.17, 40, 41 Collectively, our findings stress the need for the development of intervention strategies, including but not limited to endurance exercise training performed at a sufficient frequency and intensity for children with the ability to voluntarily activate the muscle.42
The results of the current investigation should be viewed in light of its limitations. Skeletal muscle samples from CP were collected from children and adolescents with a fixed contracture of the elbow flexors in most cases. Thus, our findings related to mitochondrial abundance and capacity in skeletal muscle from individuals with CP could be either due to the pathophysiology of the contracture per se or a more generalized feature of skeletal muscle in CP. We also recognize the fact that our study focuses on just one muscle which is a limitation for generalizability. Skeletal muscle samples from the typically developing comparison group were obtained post-mortem and represent a cumulative sample of children and adolescents after an accidental death. Thus, there was no age matching between CP and typically developing children. It should also be noted that we report measures of mtDNA/genomic DNA and OXPHOS protein abundance in CP versus typically developing muscle. For a more complete picture, ideally mitochondrial function should also have been measured. Moreover, the publicly available microarray data sets are not sampled from the biceps muscle, and this should be considered when comparing our biochemical data on biceps muscle samples with the gene network analysis on CP muscle (hamstrings muscle), aging, and bed rest (vastus lateralis muscle).
In conclusion, we observed evidence of reduced mtDNA and OXPHOS protein content in CP skeletal muscle, indicating reduced mitochondrial abundance. Moreover, transcriptional similarities to aging and long-term unloading suggests signs of metabolic alterations in CP skeletal muscle. Altogether, these aberrations in children with CP potentially influence future metabolic health of individuals with CP and tentatively contributes to the risk for metabolic diseases such as type 2 diabetes.
Acknowledgments
This work was supported by grants from the Swedish Research Council for Sports Science (FvW), the Swedish Society of Medical Research (FvW), Linnea and Josef Carlsson (RFG, FvW), Sunnerdahls handikappstiftelse (FvW, EP) and Norrbacka Eugenia Stiftelsen (FvW, EP).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.