Retrospective genetic monitoring of the threatened Yellow marsh saxifrage (Saxifraga hirculus) reveals genetic erosion but provides valuable insights for conservation strategies
Abstract
Aim
Retrospective genetic monitoring, comparing genetic diversity of extant populations with historical samples, can provide valuable and often unique insights into evolutionary processes informing conservation strategies. The Yellow marsh saxifrage (Saxifraga hirculus) is listed as ‘critically endangered’ in Ireland with only two extant populations. We quantified genetic changes over time and identified genotypes in extant populations that could be used as founders for reintroductions to sites where the species is extinct.
Location
Ireland.
Methods
Samples were obtained from both locations where the species is currently found, including the most threatened site at the Garron Plateau, Co. Antrim, which held only 13 individuals during 2011. Herbarium samples covering the period from 1886 to 1957 were obtained including plants from the same area as the most threatened population, as well as three extinct populations. In total, 422 individuals (319 present-day and 103 historical) were genotyped at six microsatellite loci. Species distribution modelling was used to identify areas of potentially suitable habitat for reintroductions.
Results
Level of phenotypic diversity within the most threatened population was significantly lower in the present-day compared with historical samples but levels of observed heterozygosity and number of alleles, whilst reduced, did not differ significantly. However, Bayesian clustering analysis suggested gradual lineage replacement over time. All three measures of genetic diversity were generally lower at the most threatened population compared with the more substantial extant populations in Co. Mayo. Species distribution modelling suggested that habitat at one site where the species is extinct may be suitable for reintroduction.
Main conclusions
The dominant genetic lineage in the most threatened population is rare elsewhere; thus, care needs to be taken when formulating any potential reintroduction programme. Our findings highlight both the need for genetic monitoring of threatened populations, but also for its swift implementation before levels of diversity become critically low.
Introduction
Knowledge of levels and patterns of intraspecific genetic diversity represents a fundamental aspect of modern conservation biology. Researchers and policy makers are now aware of the various implications of habitat loss and population extinction on diversity below the species level. Such information is crucial in the estimation of effective and minimum viable population sizes, as well as levels of inbreeding and adaptive potential (Allendorf & Luikart, 2007; Schwartz et al., 2007). These factors are particularly relevant in populations comprising very low numbers of individuals and typically those in immediate need of conservation.
Whilst there have been several recent attempts to predict the potential impacts of future habitat loss and/or population extinction on species' genetic diversity (Bálint et al., 2011; Beatty & Provan, 2011; Provan & Maggs, 2012), relatively few studies have directly quantified historical loss of diversity due to past (and ongoing) extinctions. These studies generally rely on the genetic analysis of museum or herbarium samples and have often highlighted loss of genetic diversity in extant populations compared with their historical counterparts (reviewed in Wandeler et al., 2007 and Leonard, 2008). Such retrospective genetic monitoring can provide valuable, and often unique, insights into evolutionary processes that can inform future conservation programmes (Schwartz et al., 2007; Wandeler et al., 2007; Jackson et al., 2012).
The Yellow Marsh Saxifrage (Saxifraga hirculus) is a perennial herbaceous plant with a circumpolar distribution (Hedberg, 1992). The species originated in central Asia (Hedberg, 1992), but the sole phylogeographic study carried out to date identified Alaska as the centre of genetic diversity, suggesting survival in an Alaskan/Beringian refugium during the Pleistocene glaciations (Oliver et al., 2006). The species suffered dramatic declines throughout in Europe during the last c. 200 years, primarily as a result of habitat loss, principally wetlands, with remaining populations being small and widely scattered (Vittoz et al., 2006). The Irish Red Data Book for Vascular Plants lists the species under the International Union for the Conservation of Nature (IUCN) category of ‘critically endangered’ (Curtis & McGough, 1988). We analysed herbarium samples spanning over 150 years, including extinct populations, as well as samples representing populations from the only two locations where the species is currently extant, to determine whether there has been loss of genetic variation and to use this information to formulate species augmentation and reintroduction programmes further informed by species distribution modelling, which was used to identify potentially suitable habitat for any reintroductions.
Methods
Study sites
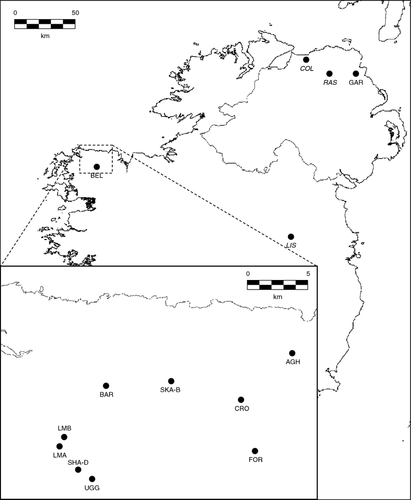
Only two populations of S. hirculus occur in Ireland; the most threatened population consisted of only 13 individuals at the Garron Plateau, County Antrim during 2011 whilst the other is substantially larger occurring at 13 sites (each with c. 100–200 individuals) near Bellacorcik, Co. Mayo. Herbarium samples show that the species occurred at much greater abundance at the Garron Plateau in the past and also occurred at another site near Rasharkin, County Antrim. Two further populations, one near Coleraine, County Derry and one near Lisclogher Co. Westmeath are now extinct.
Study species
Chromosome numbers for S. hirculus include 2n = 16, 2n = 24 and 2n = 32, although all populations outside the Arctic Circle studied to date, including those in Europe, are tetraploid (2n = 32). Reproduction is both sexual and asexual. Flowers are markedly protandrous, but self-compatible, and are pollinated by a wide range of insects, with different pollinators in different parts of the species' range (Olesen & Warncke, 1989; Warncke et al., 1993). Seeds lack special adaptations for dispersal and are generally deposited close to the parent plant (Olesen & Warncke, 1989). Vegetative reproduction occurs via rhizomes and is believed to be an important mechanism of propagation (Olesen & Warncke, 1990).
Sampling and DNA extraction
A total of 319 present-day samples were obtained during 2011. All 13 plants from the Garron Plateau, Co. Antrim were sampled, and 306 samples were collected from all 13 sites included within the Co. Mayo population (24 individuals from each with the exception of site SHE-D, for which 18 individuals were analysed). The species is protected under the Wildlife Acts 1976–2010 (Ireland) and Schedule 8 of the Wildlife (Northern Ireland) Order (1985), and it was an offence to pick, uproot or destroy the plant. Consequently, a single leaf was taken from each plant under Government licence. Samples were stored in silica gel for transportation. A total of 103 historical samples were obtained from individual plants on herbarium sheets representing both extant locations in Counties Antrim and Mayo, as well as from extinct populations at Rasharkin, Co. Antrim, Coleraine, Co. Derry and Lisclogher, Co. Westmeath. The sampling regime of herbarium samples and their distribution are given in Table 1 and Figure 1 (for herbarium codes see Table S1).
| County | Population | Year | Code | Lat | Long | N | A a | H O a | H a |
|---|---|---|---|---|---|---|---|---|---|
| Co. Antrim | Garron Plateau | 1886 | GAR-1886 | 54.990 | −6.096 | 5 | 18 | 0.600 | 0.768 |
| 1889 | GAR-1889 | 2 | N/A | N/A | N/A | ||||
| 1914 | GAR-1914 | 3 | N/A | N/A | N/A | ||||
| 1920 | GAR-1920 | 2 | N/A | N/A | N/A | ||||
| 1922 | GAR-1922 | 2 | N/A | N/A | N/A | ||||
| 1955 | GAR-1955 | 8 | 23 | 0.417 | 0.649 | ||||
| 1957 | GAR-1957 | 2 | N/A | N/A | N/A | ||||
| 2011 | GAR-2011 | 13 | 19 | 0.459 | 0.445 | ||||
| Rasharkinb | 1837 | RAS-1837 | 54.9 | −6.4 | 10 | 27 | 0.533 | 0.735 | |
| 1853 | RAS-1853 | 2 | N/A | N/A | N/A | ||||
| 1857 | RAS-1857 | 6 | 23 | 0.556 | 0.767 | ||||
| 1873 | RAS-1873 | 5 | 19 | 0.500 | 0.750 | ||||
| 1884 | RAS-1884 | 6 | 20 | 0.389 | 0.711 | ||||
| Co. Derry | Coleraineb | 1800s | COL-18XX | 55.1 | −6.7 | 7 | 20 | 0.572 | 0.698 |
| Co. Mayo | Largan Mor | 2011 | LMA-2011 | 54.140 | −9.694 | 24 | 28 | 0.743 | 0.581 |
| LMB-2011 | 54.154 | −9.686 | 24 | 27 | 0.750 | 0.643 | |||
| Sheean | 2011 | SHA-2011 | 54.118 | −9.653 | 24 | 26 | 0.660 | 0.630 | |
| SHB-2011 | 54.119 | −9.652 | 24 | 30 | 0.882 | 0.790 | |||
| SHC-2011 | 54.117 | −9.656 | 24 | 27 | 0.812 | 0.699 | |||
| SHD-2011 | 54.119 | −9.651 | 18 | 25 | 0.759 | 0.770 | |||
| Uggoll | 2011 | UGG-2011 | 54.108 | −9.644 | 24 | 25 | 0.778 | 0.512 | |
| Barroosky | 2011 | BAR-2011 | 54.195 | −9.631 | 24 | 30 | 0.785 | 0.808 | |
| Sheskin | 2011 | SKA-2011 | 54.201 | −9.562 | 24 | 23 | 0.549 | 0.673 | |
| SKB-2011 | 54.198 | −9.557 | 24 | 23 | 0.500 | 0.619 | |||
| Croaghaun | 2011 | CRO-2011 | 54.182 | −9.469 | 24 | 26 | 0.708 | 0.664 | |
| Formoyle | 2011 | FOR-2011 | 54.141 | −9.448 | 24 | 22 | 0.840 | 0.560 | |
| Aghoo | 2011 | AGH-2011 | 54.257 | −9.408 | 24 | 32 | 0.729 | 0.764 | |
| Bellacorick | 1857 | BEL-1857 | 54.2 | −9.5 | 11 | 25 | 0.576 | 0.703 | |
| 1858 | BEL-1858 | 13 | 27 | 0.615 | 0.712 | ||||
| 1965 | BEL-1965 | 3 | N/A | N/A | N/A | ||||
| 1968 | BEL-1968 | 6 | 19 | 0.750 | 0.821 | ||||
| 1970 | BEL-1970 | 3 | N/A | N/A | N/A | ||||
| Co. Westmeath | Lisclogherb | 1880 | LIS-1880 | 53.6 | −7.1 | 3 | N/A | N/A | N/A |
| 1888 | LIS-1888 | 4 | N/A | N/A | N/A |
- N, number of individuals studied; A, number of alleles; HO, observed heterozygosity; H, phenotype diversity (Gini–Simpson index).
- a Number of alleles, observed heterozygosity and phenotype diversity only calculated for samples with N ≥ 5.
- b Extinct population: Latitude/Longitude approximate.
DNA was extracted from all samples using the Qiagen DNeasy Plant Mini Kit, after an initial 8 min grinding at 30 Hz using a Retsch MM300 mixer mill. DNA was quantified visually on 1% agarose gels stained with ethidium bromide and diluted to a concentration of 50 ng μl−1 for subsequent PCR. All DNA extractions from herbarium samples were carried out in a laboratory where no previous S. hirculus work had been performed.
Microsatellite genotyping
All samples were genotyped for six microsatellite markers developed for S. hirculus using the ISSR cloning method outlined in Provan & Wilson (2007). Because of difficulties associated with amplifying longer fragments from herbarium samples, primers were designed to amplify products of less than c. 200 bp (Table 2). The polyploid nature of S. hirculus in Ireland represented a further problem in scoring allele sizes accurately where stutter bands were present. Consequently, we were limited to using two trinucleotide microsatellites and two tetranucleotides, which generally display minimal stuttering, and two dinucleotides with relatively low stuttering. Forward primers were modified by the addition of a 19 bp M13 tail (5′-CACGACGTTGTAAAACGAC-3′) and reverse primers were modified by the addition of a 7-bp tail (5′-GTGTCTT-3′). PCR was carried out in a total volume of 10 μl containing 100 ng genomic DNA, 10 pmol of HEX-labelled M13 primer, 1 pmol of M13-tailed forward primer, 10 pmol reverse primer, 1× PCR buffer, 200 μm each dNTP, 2.5 mm MgCl2 and 0.25 U GoTaq Flexi DNA polymerase (Promega, Sunnyvale, CA, USA). PCR was carried out on a MWG Primus thermal cycler using the following conditions for all loci with the exception of SH-3-B11: initial denaturation at 94 °C for 3 min followed by 10 touchdown cycles of denaturation at 94 °C for 30 s, annealing at 68 °C for 30 s (−1 °C per cycle), extension at 72 °C for 30 s followed by 30 cycles (40 for herbarium samples) of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 30 s and a final extension at 72 °C for 5 min. For locus SH-3-B11, the following conditions were used: initial denaturation at 94 °C for 3 min followed by 40 cycles (50 for herbarium samples) of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s, extension at 72 °C for 30 s and a final extension at 72 °C for 5 min. Blank negative controls were routinely used, and c. 25% of herbarium samples were genotyped twice to check for artefacts resulting from low DNA quality, which were not observed. Genotyping was carried out on an AB3730xl capillary genotyping system. Allele sizes were scored using LIZ-500 size standards and were checked by comparison with previously sized control samples.
| Locus | Repeat | Primers | A | Allele size range |
|---|---|---|---|---|
| SH-1-B08 | (AGC)5 | CCCGCCATTTCTCTATACCA | 7 | 119–137 |
| GGTTGAGCCAGTCCAAGAAG | ||||
| SH-2-D03 | (CTA)5 | GCTTTTCCATTTTTAGGGCTTT | 10 | 139–169 |
| AAAAGGAAAGTGAGATACTAATTAGAACAG | ||||
| SH-3-A03 | (AT)6 | TCAAAATATTATTAAGGGAAAAATTCTCA | 8 | 156–188 |
| CCAAATGTTTGAGTTATGTATAGTTACG | ||||
| SH-3-B11 | (TCTT)7 | TGGCTACTACAATGTAAAGTTGTCTC | 8 | 132–160 |
| CATAAGTCAAAAGTCAAGGTGTCG | ||||
| SH-4-E03 | (AAAT)4 | TGTCTGTTTGGACATTCCCTTA | 11 | 136–208 |
| TCAATATATTCTTAAGTTGATTATTAAGTGTG | ||||
| SH-4-F10 | (TA)6 | GGATCCCTCACTTGAAGCTC | 16 | 122–160 |
| TGTATAGATCAACTCTGCCAAAAA |
- A, number of alleles.
- Forward tailed with CACGACGTTGTAAAACGAC.
- Reverse tailed with GTGTCTT.
Genetic analysis
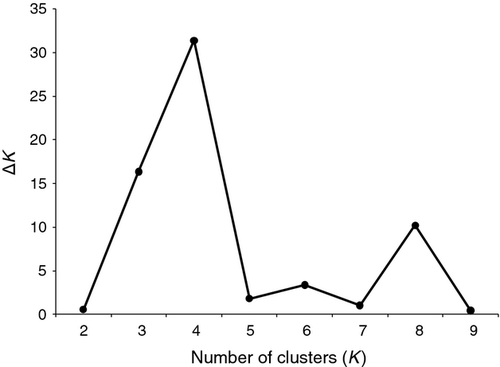
No chromosome counts are available for Irish S. hirculus, but the species is believed to be polyploid in the majority of its non-Arctic range (Hedberg, 1992), and more than two bands were observed at all six loci analysed, suggesting polyploidy. Thus, it was not possible to score genotypes based on allele frequencies, and as a result we could not carry out many standard population genetic analyses (e.g. calculation of allelic richness, AMOVA). Consequently, the patterns of alleles observed for each locus in an individual were scored as phenotypes. Within-population phenotype diversity was estimated for samples with N ≥ 5 using the total number of alleles, observed heterozygosity (HO), namely the proportion of observed heterozygous individuals in a population, and the Gini–Simpson diversity index, analogous to Nei's gene diversity, averaged over loci (Jost, 2006). Genetic clustering of individuals was assessed using a Bayesian procedure implemented in the Structure software package (V2.3.3; Pritchard et al., 2000), which can accommodate ambiguous co-dominant markers for polyploid species. The programme was run using no prior knowledge and the admixture ancestry model. Five independent runs were carried out for each value of K, the number of genetic clusters, up to K = 10, because log-likelihood values reached a peak at K = 8 and decreased thereafter. Each Markov chain Monte Carlo analysis used a burn-in period of 10,000 followed by a further 100,000 iterations. The most likely value for K was estimated using the ΔK statistic of Evanno et al. (2005) implemented in the Structure Harvester software package (V0.6.1; Earl & vonHoldt, 2012).
Species distribution modelling
A presence-only maximum entropy approach was used to predict landscape suitability for the S. hirculus throughout Ireland, from a sample set of known occurrences and spatially explicit environmental parameters. Maximum entropy has been shown to frequently outperform other presence-only modelling techniques particularly at very low sample sizes (Elith & Graham, 2009). Environmental parameters were described at a 500-m cell resolution (Table S2). The software package MaxEnt was used (V3.3.3k; Phillips et al., 2006). To maximize model flexibility, we considered linear, quadratic, threshold and hinged functions for all environmental parameters (Phillips & Dudík, 2008). Due to the paucity of records, it was not possible to segregate the data set into a training and test sets, thus only a training set was used (Farren et al., 2010). Jackknife resampling analysis was used to determine a heuristic estimate of the relative contribution of each variable based on the performance of the global model (known as the regularized gain) without the variable of interest compared with the influence of that variable in isolation (derived from a univariate model only). Global model performance was judged using the area under the receiver operating characteristic (ROC) curve (Liu et al., 2005). Marginal response curves of the predicted probability of species occurrence were graphed for each explanatory variable. A map of landscape favourability was generated using ArcGIS 9.3 (ESRI, California, USA), and the 10th percentile training presence was used as the threshold.
Results
Between seven and 16 alleles were detected across the six loci analysed (average 10 alleles per locus; Table 2). The total number of alleles per population with sample numbers N ≥ 5 ranged from 18 (GAR-1886 population) to 32 (AGH-2011 population). Observed heterozygosity (HO) ranged from 0.389 (RAS-1884 population) to 0.882 (SHB-2011 population; Table 1). Levels of phenotype diversity (H) ranged from 0.387 (GAR-2011 population) to 0.821 (BEL-1968 population; Table 1). The extant Garron Plateau population had the third lowest level of HO, after the GAR-1955 sample, and the lowest level of H. Both values (0.436 and 0.387, respectively) were lower than those in the extant Co. Mayo population, where HO ranged from 0.500 to 0.882 (mean = 0.730) and H ranged from 0.512 to 0.808 (mean = 0.670). For the Garron Plateau population, the level of H (calculated over all individuals, i.e. 24 individuals sampled across six time points vs. 13 samples from 2011) was significantly higher in the herbarium samples than in the extant samples, whereas levels of HO and average number of alleles per individual (A), whilst higher for herbarium samples (again calculated over all individuals), were not significantly different (Table 3).
| Period | Diversity | ||
|---|---|---|---|
| A | Ho | H | |
| Historical (pre-1958) | 1.732 | 0.507 | 0.710 |
| Extant | 1.722 | 0.459 | 0.445 |
| NS | NS | P = 0.013 | |
- A, mean number of alleles; HO, mean observed heterozygosity; H, mean phenotype diversity (Gini–Simpson index).
- Significance of differences in mean values was estimated using a t-test.
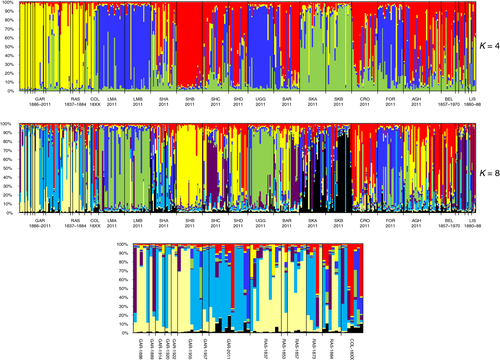
The results of the Bayesian clustering analysis indicated that the most likely number of genetic clusters was K = 4, followed by K = 8 (Fig. 2). For K = 4 genetic clusters (Fig. 3, top), the vast majority of plants from the Garron Plateau and the now extinct Rasharkin population were predominantly associated with the cluster shown in yellow, which was comparatively rare elsewhere in Ireland. The population from Co. Mayo displayed varying degrees of admixture, from being almost completely dominated by a single cluster (red at SHB) to being comprised of a substantial fraction of all four clusters (SHA). Assignment of populations to K = 8 clusters (Fig. 3, middle and bottom) revealed some further subtle genetic substructuring, particularly in the Northern Ireland populations. This shows a distinct temporal shift from the lineage represented by light yellow, which was initially the dominant lineage at both the Garron Plateau and Rasharkin populations, to the lineage represented in light blue, to the point where the light yellow lineage is now almost completely absent.
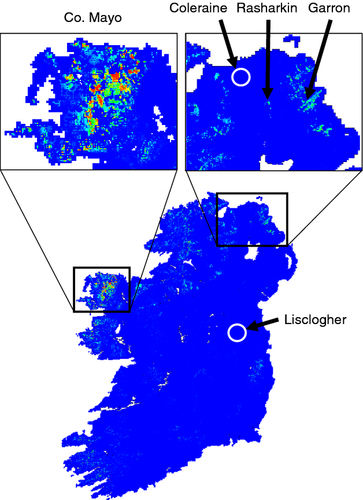
Saxifraga hirculus occurrence was strongly associated with landscapes dominated by bog, fen, marsh and swamp typically on high-altitude (> 175 m above sea level) plateaus (i.e. low hilliness index), which experienced low maximum temperatures (< 15 °C), high precipitation (1000–1700 mm of rain annually) and low seasonality, that is, consistently cool and wet all year round (Figures S1 and S2). These regions were typically negatively correlated with agricultural pastures. Model performance, defined as the area under the curve or AUC = 0.998. The model suggested that the extent of suitable habitats for S. hirculus is limited (Fig. 4a). Highest suitability and the greatest extent of habitat was predicted throughout north Co. Mayo (Fig. 4b). The model also suggested that less optimal habitat was found throughout Co. Antrim including the Garron Plateau (Fig. 4c). A patch of potentially suitable habitat (determined using the 10th percentile training presence = 0.692) was also identified near Rasharkin, which may represent the site of an extinct population. The habitat at locations of other now extinct populations near Coleraine, Co. Derry and Lisclogher, Co. Westmeath was predicted to have vanished.
Discussion
Saxifraga hirculus genetic erosion over time
Maintaining genetic variation in threatened populations, which by their nature tend to be small and/or fragmented, is one of the central tenets of conservation genetics (Allendorf & Luikart, 2007; Schwartz et al., 2007). The retrospective genetic monitoring afforded by the comparison of a critically threatened extant population with historical samples from the same area suggests that levels of population genetic diversity of S. hirculus in the critically endangered Garron Plateau, Co. Antrim are not only far lower than those in the other, larger extant populations in Co. Mayo, but also appreciably lower than the average values from historical samples (pre-1958) from the same area, although there is evidence of some fluctuation in these values through time. This loss of genetic diversity has been accompanied by the replacement of one lineage identified by the Bayesian clustering analysis (shown in Fig. 2, bottom, in light blue) by another (shown in light yellow), to the extent that the new lineage accounts for over 60% of the total genetic diversity. These changes are probably the result of a combination of factors, foremost among which are stochastic fluctuations in allele frequencies due to the greatly exaggerated effects of genetic drift in very small populations. These changes may also have been accompanied by a founder effect, because the Garron Plataeu population was recorded as extinct after 1920, but subsequently ‘refound’ in 1955 (Kertland, 1956). The extinction of the population at Rasharkin, Co. Antrim at the end of the 19th century would have contributed further to the loss of this lineage, both directly and indirectly via the cessation of possible gene flow between the two Co. Antrim populations. Nevertheless, although the levels of phenotypic diversity were significantly lower in the extant population than in the historical samples, the number of alleles was not. Given that the number of alleles is correlated with the ability to respond to selection, this does not appear to be as a serious concern as potential inbreeding depression.
Conservation implications
Although information from population genetic studies can inform best-practice conservation strategies, the long decline in S. hirculus population numbers highlights a further conservation dilemma. Numbers of recorded individuals at the Garron Plateau have fallen from 130 during 1999 to 13 during 2011 (Georgina Thurgate, pers. comm.). Despite the importance of vegetative reproduction in the spread and persistence of S. hirculus (Olesen & Warncke, 1990), we detected only two potentially clonal individuals (i.e. 15% shared identical multilocus phenotypes). Nevertheless, the extremely small number of individuals comprising the extant population at the Garron Plateau leaves it extremely vulnerable to sudden, stochastic extinction. Thus, it is clear that some sort of augmentation programme is necessary. One of the goals of conservation genetics is to ensure that the provenance of individuals used for such programmes is closely aligned with the population (Lesica & Allendorf, 1999), but the Garron Plateau individuals are generally associated with a genetically distinct group that is not found at any significant level elsewhere in the remaining populations in Ireland. Re-establishment or augmentation of a population using genetically divergent individuals results in a trade-off between increasing population numbers at the risk of outbreeding depression (Edmands, 2007). The Bayesian clustering analysis suggests that if a source population is required for augmentation, it should be the SHA population in Co. Mayo, which not only has the highest percentage assignment to the predominant cluster found in the Garron Plateau, but also contains a mixture of several different lineages. This represents a further important aspect of strategic conservation based on molecular genetic approaches, namely the fact that data from a low number of putatively neutral loci do not necessarily provide insights into the relative fitness of genotypes, particularly in differing habitats (Ennos et al., 1997; Hollingsworth et al., 1998). Augmentation or introduction of a range of genotypes, including those closest to genotypes found in the extant Garron population, as is the case for the Co. Mayo SHA population, will provide a balance between ‘genotype matching’ and sufficient variation for natural selection to operate on. Of course, reintroduction need not be limited to material from a single source population, and the results of the genetic clustering analysis could be used to identify the widest possible range of genetic diversity for reintroduction, if so desired.
Saxifraga hirculus occurrence was associated with bog, fen, marsh and swamp typically on high-altitude plateaus with low maximum temperatures, high precipitation and low seasonality, that is, consistently cool and wet. The loss of the Coleraine, Co. Derry and Lisclogher, Co. Westmeath populations could be attributed to loss of such habitat, because the species distribution model did not identify these areas as currently suitable. A number of areas in the Garron Plateau, Co. Antrim were identified as potentially suitable for establishing new populations (via ex situ conservation) to further supplement and expand the extant population. A single area of potentially suitable habitat was identified near the site of the now extinct population at Rasharkin, Co. Antrim. Insights into the historical genetic makeup of the now extinct Rasharkin population afforded by the analysis of the herbarium samples mean that a controlled reintroduction programme, based on recreating the genetic make-up of the original population from individuals extant elsewhere (i.e. Co. Mayo), could maximize the potential success of re-establishing this lost population.
The genetic changes revealed by the retrospective genetic monitoring indicate the need to implement such approaches as soon as possible. Regular censuses of the population at the Garron Plateau began during 1999 when there were 130 plants. If genetic monitoring had commenced at the same time there would have been more chance of developing a successful ex situ conservation programme to maximize genetic diversity than at present. As it is, the current scenario further highlights the need for conservation practitioners to move away from a ‘fire-fighting’ mentality (Mace & Purvis, 2008). Nevertheless, the findings of our study can be used to inform any potential reintroduction/augmentation programmes. Results from a previous re-establishment programme in Scotland indicate that ex situ propagation of seedlings followed by transplantation is a more successful method than simply sowing seeds directly onto potential recovery sites (Welch, 2002). Based on the information from the current study, genetic analysis of ex situ individuals could be used to select individuals most representative of the current extant gene pool, whilst aiming to maximize genetic diversity.
Acknowledgements
This study was commissioned and funded by the Natural Heritage Research Partnership between the Northern Ireland Environment Agency (NIEA) and Quercus, Queen's University Belfast. We are grateful to Kyle Hunter and Mark Wright (NIEA) and James Kilroy, Cameron Clothworthy, Leonard Floyd and Deirdre Lynn from the National Parks & Wildlife Service, Department of Arts, Heritage and the Gaeltacht, Republic of Ireland for sample collection. Herbarium samples were provided by Colin Kelleher, Noeleen Smyth and Mathew Jebb (National Botanic Garden of Ireland) and Peter Crowther (Ulster Museum, National Museums Northern Ireland). We are also grateful to Associate Editor Andy Lowe and two anonymous referees for constructive comments that greatly improved the manuscript. Many thanks to Georgina Thurgate who acted as NIEA Client Officer.
References
Biosketches
Gemma Beatty is a Post-doctoral Research Fellow at Queen's University Belfast. Her PhD research compared how post-glacial recolonization and range-edge effects have shaped the genetic diversity of several Monotropoideae species. She is interested in using genetic approaches to study the effects of past and present climate change on the distribution ranges of natural populations, and the various factors that determine these ranges.
Neil Reid is Manager of Quercus, Northern Ireland's Centre for Biodiversity and Conservation Biology and has a background in species distribution modelling as a tool for identifying high conservation value areas for endangered species.
Jim Provan is a Reader in Evolutionary Genetics at Queen's University Belfast. His research interests focus on how genetic variation is distributed across species ranges, and on the effects of past, present and future climate change on levels and patterns of intraspecific diversity.
Author contributions: N.R. and J.P. conceived the study; G.E.B. and J.P. collected the data; All three authors carried out the analyses and wrote the manuscript.




