Merkel cell polyomavirus is a passenger virus in both poroma and porocarcinoma
Funding information: Helsinki University Central Hospital, Grant/Award Number: TYH2020208
Abstract
Background
Merkel cell polyomavirus (MCPyV) has been studied in several malignant and nonmalignant tissues. However, only in Merkel cell carcinoma (MCC) has the connection to tumorigenesis been established. Previously, eccrine porocarcinoma samples were shown to express MCPyV in the majority of samples. We aimed to examine MCPyV in porocarcinoma and poroma samples using MCC as the reference material.
Methods
We analyzed 17 porocarcinoma and 50 poroma samples for the presence of MCPyV using LT antigen immunostaining and DNA detection methods. In addition, 180 MCC samples served as controls.
Results
MCPyV LT antigen immunostaining was detected in 10% of poroma and 18% of porocarcinoma samples; on the other hand, it was present in 65% of MCC samples. MCPyV DNA was detected in only 10% of poroma and porocarcinoma samples compared with 96% of MCC samples. The viral DNA copy number in all MCPyV DNA-positive MCCs was at least 25 times higher than that in porocarcinoma or poroma samples with the highest MCPyV DNA-to-PTPRG ratio.
Conclusions
The low number of viral DNA copies in poroma and porocarcinoma samples, together with the negative LT expression of MCPyV DNA-positive tumors, indicates that MCPyV is simply a passenger virus rather than an oncogenic driver of porocarcinoma.
1 INTRODUCTION
Eccrine porocarcinoma (EPC) is a rare eccrine sweat gland-originating skin carcinoma. EPC occurs predominantly in the elderly, with a mean age at diagnosis varying from 62 to 76 years.1, 2 In our recent study, the age-adjusted prevalence of EPC was 1.5-fold higher in men than in women in Finland: 0.06 for men and 0.04 for women per 100 000 person-years using World Standard Population, Figure 2. EPCs typically occur in the head and neck area and in the lower limbs.1
In previous reports, the course of EPC was described as aggressive, although recent studies suggest that an aggressive behavior and progression to metastasis may represent exceptions.3-5 Rather unusually, EPC's predecessor, poroma, is a benign adnexal neoplasm: No consensus exists whether all or only a minority of poromas progress to EPCs.6 Knowledge of the tumorigenesis in EPC remains limited, while some studies indicate that immunosuppressive conditions and opportunistic infections may contribute to cancer progression given that EPC disproportionally affects organ transplant recipients and patients with hematological malignancies.7, 8
The pathogenesis of Merkel cell carcinoma (MCC) is driven by the duality of Merkel cell polyomavirus (MCPyV) infection and chronic exposure to ultraviolet radiation. MCPyV plays a causative role in the oncogenesis of MCC in most cases. The MCPyV genome is clonally integrated into MCC tumor cells, whereby MCPyV infection and integration occur before clonal expansion of the tumor cells.9 In addition to the clonal integration of the virus genome, T-antigen truncating mutations and the expression of viral oncoproteins named T antigens are strictly associated with specific MCC oncogenic events.9 Immunohistochemistry and a mouse monoclonal CM2B4 antibody are reliable and suitable methods for routine clinical use to detect MCPyV-positive MCCs.10
Since the discovery of MCPyV, numerous normal, premalignant, and malignant tissues have been scrutinized for MCPyV.11 Traces of MCPyV genomic material are detected in both noncancerous and cancerous tissues, indicating its status as a passenger virus with no causal role.12, 13 MCC studies on MCPyV frequently showed that a viral copy number per cell or viral gene-to-human control gene ratio determined by quantitative PCR typically reaches at least 0.01 to 0.1 or greater when a viral large T (LT) antigen expression is detected in tumor cells, thereby representing a generally acceptable level for true MCPyV positivity.14
In this current study, we sought to examine MCPyV in poromas and EPC samples, using MCC samples as the reference material.
2 MATERIALS AND METHODS
The Ethics Committee of Helsinki University Hospital and the local review board approved the study plan and protocol. Following approval from the Helsinki University Hospital, we sent a query to Finnish biobanks requesting EPC and poroma tumor samples with the corresponding clinical data. After verification of the existing tumor samples, we sent requests for samples to the Helsinki Biobank (Helsinki Finland), Finnish Clinical Biobank (Tampere, Finland), and Biobank Borealis (Oulu Finland). Once individual approval was received, tissue samples were collected.
For MCC samples, the Ministry of Health and Social Affairs granted us permission to collect patient data, while the National Authority for Medicolegal Affairs approved our request to collect and analyze the tissue samples.
2.1 Tumor samples
Altogether, 17 porocarcinoma and 50 poroma samples were available for analysis. In addition, we analyzed 180 MCC samples.
2.2 Immunohistochemistry
Tissue microarrays were constructed from poroma and porocarcinoma tissue samples by using TMA Grand Master tissue microarrayer. Four-micrometer sections were cut on Labsolute microscope slides (Art. Nr. 7695015, The Geyer & Co, KG, Germany) deparaffinized, rehydrated, and endogenous peroxidase was blocked with 0.7% hydrogen peroxide. An epitope retrieval was carried out in sodium citrate (pH 6.0, 10 mmol/L) in a water bath (at 98°C for 20 minutes). LT antigen expression of MCPyV was detected by using mouse monoclonal antibody (clone CM2B4, sc-136 172, Santa Cruz Biotechnology Inc, Texas). Primary antibody was diluted on Normal Antibody Diluent (dilution 1:100; ImmunoLogic a WellMed Company, the Netherlands) and the mixture was incubated on slides for 1 hour at a room temperature. The primary antibody was detected using a BrightVision Poly-HRP anti-mouse IgG histostaining kit (ImmunoLogic) and ImmPACT DAB Substrate (Vector Laboratories, California) according to manufacturers' instructions.
2.3 MCPyV DNA detection
Tissue sections were available for the DNA extraction in 119 MCC and in 67 EPC or poroma samples. Three tumor tissue representative areas of 1.0-μm diameter were punched from formalin-fixed paraffin-embedded tissue blocks using TMA Grand Master tissue microarrayer (3DHISTECH Ltd, Hungary). A genomic DNA was extracted using QIAsymphony DSP DNA mini kit and QIAsymphony SP instrument (Qiagen GmbH, Germany) according to manufacturer's instructions. MCPyV DNA-to-reference gene (protein tyrosine phosphatase gamma receptor, PTPRG) ratio was investigated using quantitative PCR as described in detail elsewhere.15 Shortly, the DNA was amplified in a 20 μL reaction PCR mixture consisting of Probe Master reagents and fluorescein-labeled locked nucleic acid probes (Roche Diagnostics GmbH, Germany) using CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Inc, California). The relative MCPyV DNA-to-PTPRG gene ratio for each tissue sample was calculated using delta-delta Ct method.
3 RESULTS
3.1 Poroma and eccrine porocarcinoma samples
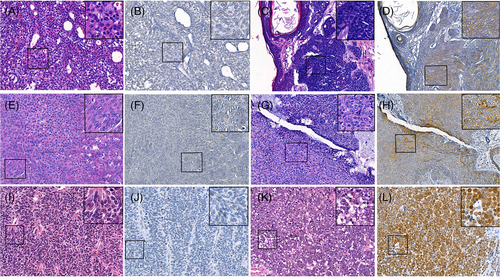
Weak-to-strong heterogeneous MCPyV LT antigen immunostaining was detected in five of 49 (10.2%) poroma and three of 17 (17.6%) EPC tissues samples (Figure 1, Table 1).
| LT antigen expression | MCPyV DNA | P | |
|---|---|---|---|
| Negative, n (%) | Positive, n (%) | ||
| Merkel cell carcinoma | |||
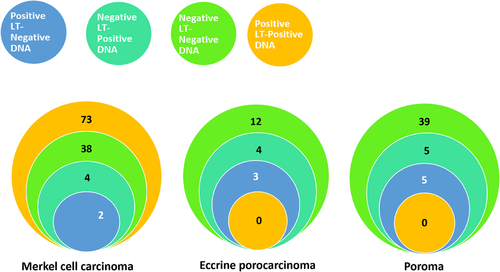
| Negative | 38 (90.5) | 4 (9.5) | <.0001 |
| Positive | 2 (2.7) | 73 (97.3) | |
| Not available | 2 | ||
| Porocarcinoma | |||
| Negative | 12 (85.7) | 2 (14.3) | >.5 |
| Positive | 3 (100) | 0 (0) | |
| Poroma | |||
| Negative | 39 (88.6) | 5 (11.4) | >.5 |
| Positive | 5 (100) | 0 (0) | |
MCPyV DNA was detected in only seven of 67 (10.4%) poroma or porocarcinoma samples, Figure 2. The relative MCPyV DNA-to-PTPRG gene ratio for virus-positive samples varied from 0.0001 to 0.0156 (median = 0.0004). None of the tumors containing MCPyV DNA were positive for LT expression in the immunohistochemistry. The presence of MCPyV DNA or LT expression did not correlate with age at diagnosis, tumor size, sex, or location in poroma or porocarcinoma, all P values >.05, Table 2.
| Porocarcinoma | Poroma | |||||||
|---|---|---|---|---|---|---|---|---|
| LT expression | MCPyV DNA | LT expression | MCPyV DNA | |||||
| Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | |
| Sex | ||||||||
| Female | 7 (87.5) | 1 (12.5) | 7 (87.5) | 1 (12.5) | 25 (92.6) | 2 (7.4) | 25 (92.6) | 2 (7.4) |
| Male | 7 (77.8) | 2 (22.8) | 8 (88.9) | 1 (11.1) | 17 (85.0) | 3 (15.0) | 17 (85.0) | 3 (15.0) |
| N.A. | 2 | 2 | ||||||
| Location | ||||||||
| Head | 4 (80.0) | 1 (20.0) | 5 (100) | 0 (0) | 9 (90.0) | 1 (10.0) | 8 (72.7) | 3 (27.3) |
| Limb | 5 (83.3) | 1 (16.7) | 5 (83.3) | 1 (16.7) | 22 (91.7) | 2 (8.3) | 21 (91.3) | 2 (8.7) |
| Trunk | 5 (83.3) | 1 (16.7) | 5 (83.3) | 1 (16.7) | 12 (92.3) | 1 (7.7) | 13 (100) | 0 (0) |
| N.A. | 1 | 1 | 2 | |||||
| Age at diagnosis | ||||||||
| Median (range) | 73 (19-89) | 72 (22-74) | 72 (19-89) | 72.5 (71-74) | 65 (7-89) | 61 (35-75) | 65 (7-89) | 62 (59-75) |
| Size (mm) | ||||||||
| Median (range) | 22.5 (5-150) | 30 (5-55) | 20 (5-65) | 150 | 14 (3-55) | 20 (5-35) | 14 (3-55) | 16 (5-25) |
| N.A. | 2 | 1 | 2 | 1 | 9 | 3 | 11 | 1 |
3.2 Merkel cell carcinoma samples
Tissue microarray samples were available from 166 of 180 MCC samples, for which LT expression was detected in 107 (64.5%) samples.
MCPyV DNA was frequently detected in MCC. Specifically, 114 of 119 (95.8%) samples analyzed contained detectable amounts of virus DNA. Furthermore, the MCPyV DNA-to-PTPRG gene ratio in the virus-positive tissues varied from 0.0005 to 2905 (median = 2.004).
The viral DNA copy number frequently exceeded 0.4 in the immunostaining LT-positive MCCs, which was used as the cut-off value for positivity. Only two LT-positive tumors exhibited a copy number <0.4 and 4 DNA-positive tumors showed no LT immunostaining (Table 1). The association between LT expression and the presence of viral DNA was highly statistically significant (χ2, P < .0001).
In all MCPyV DNA-positive MCCs, the virus DNA copy number was at least 25 times greater than that in EPC and poroma samples with the highest MCPyV DNA-to-PTPRG ratio (0.0156).
4 DISCUSSION
Herein, we studied the presence of MCPyV in poroma and EPC samples using both immunohistochemistry and quantitative PCR, while MCC samples served as the reference material. Only 10% of poroma samples and 18% of EPC samples showed immunopositivity for the LT antigen. Furthermore, MCPyV DNA was detected in only seven (10%) of the poroma and EPC samples. The viral DNA copy numbers were low in both poroma and EPC in comparison to MCPyV DNA-positive MCC samples. To our knowledge, only one previous case report has detected MCPyV DNA by PCR in poroma.16
The virus DNA copy number was at least 25 times higher in MCCs, indicating that MCPyV is merely a passenger virus in poromas and EPCs. Truncated LT and viral genome integration to the tumor genome represent hallmarks of MCPyV-positive MCC tumors.9 In this study, a low MCPyV copy number and the absence of LT expression in virus-positive tumors indicate that the MCPyV genome did not clonally integrate into poroma or porocarcinoma cell genomes and, thus, did not contribute to early tumorigenesis. Immunostaining for the LT antigen with the mouse monoclonal antibody CM2B4 is considered a reliable and specific method for indicating the presence of MCPyV, while the integrated MCPyV genome expresses LT in MCC.10, 15 However, like here, false-positive LT-immunostaining samples appear in MCPyV DNA-negative tissues.11
Although MCPyV has been found in numerous non-MCC tumors,11 MCPyV appears not to actively participate in the onset and maintenance of cancer growth, as evidenced by the absence of LT or given the nondetectable expression. MCPyV replication is limited to dermal fibroblasts and potentially to the lung.17 In addition, MCPyV is part of the normal human skin microbiome18 given that it is habitually shed from healthy individuals' skin. Thus, contamination may represent one reason for the low levels of MCPyV detected.18
Virus-positive MCC tumors contain >1 MCPyV genome copies per cell.12 In a previous study, Urso et al detected low copy numbers of viral DNA in EPC, comparable to the virus copy number levels in healthy skin tissue19 . This agrees with our study and other non-MCC skin tumor studies, in which, excluding MCC, only low MCPyV copy numbers are detected.20 Here, we found that LT-expressing MCCs exhibited a significantly higher MCPyV DNA copy number than MCPyV-negative MCCs, poromas, or EPC tumors. Additionally, none of the MCPyV DNA-positive EPCs or poromas expressed the LT antigen in tissues, supporting the passenger role of MCPyV in EPC.
However, the prevalence of the DNA-positive EPCs differs significantly between our study (10%) and Urso et al (68%).19 This difference might be explained by the variability in the methods applied. Specifically, we used different primer sets and quantitative PCR to detect viral DNA; on the other hand, Urso et al used droplet digital PCR, which is known to be less prone to inhibitors in tissues and better at detecting low copy number variants, while also containing a higher risk for contamination.21
Previous findings consist of a single case report from two MCC cases developing in an existing poroma.22 That report, while interesting, lacks information on the tumors' MCPyV status. Thus, causality for MCPyV in the development of these tumors remains unsubstantiated.
To conclude, our study indicates that MCPyV may not be an oncogenic driver for EPC. It must be noted that in MCPyV DNA-positive, CM2B4-negative poromas and porocarcinomas the viral copy number was too low to provide immunohistochemical expression of the LT antigen, not that it was absent from the tumor cell genome. Therefore, further studies focusing on the tumorigenesis of EPC are necessary.
ACKNOWLEDGMENTS
This study was funded by Helsinki University Central Hospital (HUCH) Competitive Research Fund (EVO) Grant number TYH2020208.
CONFLICT OF INTEREST
Each author declares no financial conflicts of interest regarding the data presented in this manuscript.
AUTHOR CONTRIBUTIONS
Virve Koljonen and Harri Sihto conceived the study idea and designed the study idea. Anna-Stiina Meriläinen and Virve Koljonen prepared first draft of the manuscript. Harri Sihto prepared the figures. All authors contributed to the writing of the manuscript. All authors contributed to proof-reading and revising of the manuscript.
ETHICS STATEMENT
The Ethics Committee of Helsinki University Hospital and the local review board approved the study plan and protocol. Following approval from the Helsinki University Hospital, we sent a query to Finnish biobanks requesting EPC and poroma tumor samples with the corresponding clinical data. After verification of the existing tumor samples, we sent requests for samples to the Helsinki Biobank (Helsinki Finland), Finnish Clinical Biobank (Tampere, Finland), and Biobank Borealis, (Oulu Finland). Once individual approval was received, tissue samples were collected. For MCC samples, the Ministry of Health and Social Affairs granted us permission to collect patient data, while the National Authority for Medicolegal Affairs approved our request to collect and analyze the tissue samples.
Open Research
DATA AVAILABILITY STATEMENT
Data available on request from the authors.




