Delineation of the clinical profile of CNOT2 haploinsufficiency and overview of the IDNADFS phenotype
Funding information: This work was supported, in part, by grants from the Italian Ministry of Health (Ricerca 5 per Mille, to M.T.), and Fondazione Bambino Gesù (Vite Coraggiose).
Abstract
CNOT2 haploinsufficiency underlies a rare neurodevelopmental disorder named Intellectual Developmental disorder with NAsal speech, Dysmorphic Facies, and variable Skeletal anomalies (IDNADFS, OMIM 618608). The condition clinically overlaps with chromosome 12q15 deletion syndrome, suggesting a major contribution of CNOT2 haploinsufficiency to the latter. CNOT2 is a member of the CCR4-NOT complex, which is a master regulator of multiple cellular processes, including gene expression, RNA deadenylation, and protein ubiquitination. To date, less than 20 pathogenic 12q15 microdeletions encompassing CNOT2, together with a single truncating variant of the gene, and two large intragenic deletions have been reported. Due to the small number of affected subjects described so far, the clinical profile of IDNADFS has not been fully delineated. Here we report five unrelated individuals, three of which carrying de novo intragenic CNOT2 variants, one presenting with a multiexon intragenic deletion, and an additional case of 12q15 microdeletion syndrome. Finally, we assess the features of IDNADFS by reviewing published and present affected individuals and reevaluate the clinical phenotype of this neurodevelopmental disorder.
1 INTRODUCTION
12q15 deletion syndrome refers to a clinically heterogeneous neurodevelopmental disorder characterized by developmental delay (DD), intellectual disability (ID), hypotonia, speech delay with nasal speech, and variable dysmorphic facial features.1 Since the first report, a few individuals with overlapping 12q15 deletions have been reported.2, 3 Studies directed to characterize the minimal critical region of 12q15 deletion syndrome have suggested CNOT2 haploinsufficiency as the driven event underlying this disorder.4, 5 The CCR4-NOT transcription complex subunit 2 (CNOT2, MIM*604909) gene encodes a component of the widely expressed CCR4-NOT complex, which is implicated in various cellular processes, including mRNA degradation, miRNA-mediated repression, translational repression during translational initiation, thus behaving as a master regulator of gene expression during embryonic development.6-8 To date, less than 20 pathogenic structural rearrangements encompassing CNOT2, either deletions or duplications, have been reported in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar). Among these, 13 are microdeletions. More recently, two intragenic CNOT2 microdeletions and a truncating gene variant have sporadically been highlighted as a new cause of a developmental disorder clinically related to 12q15 deletion syndrome.9-11 Due to this clinical overlap, the disorder has been renamed with the acronym IDNADFS (Intellectual Developmental disorder with NAsal speech, Dysmorphic Facies, and variable Skeletal anomalies, MIM*618608), which also includes 12q15 deletion syndrome. However, the clinical profile of IDNADFS appears variable and has not fully been delineated yet, and its relationship with 12q15 deletion syndrome is still debated.5 Indeed, due to scarce reports describing affected individuals, it remains unclear whether the full-blown clinical profile of this disorder can exclusively be ascribed to CNOT2 haploinsufficiency or whether other genes included in the deleted region (i.e., CCT2, FRS2, CPSF6, PTPRB, and ZFC3H1) may contribute to the phenotypic variability.5
Here we report five unrelated individuals carrying intragenic CNOT2 variants, a multiexon intragenic deletion, and a 7.3 Mb deletion encompassing the gene. By reviewing the clinical features of the affected subjects with intragenic CNOT2 rearrangements/variants from previous reports and the present series, we also reassessed the clinical phenotype of IDNADFS, and its overlap with the 12q15 deletion syndrome.
2 SUBJECTS AND METHODS
2.1 Study design and approval
Five individuals affected by an unrecognized condition severely impairing neurodevelopment and then found to have molecular events affecting CNOT2 were enrolled in the context of the present research project. All clinical investigations were conducted according to the declaration of Helsinki. The project was approved by the local Institutional Ethical Committee of the Ospedale Pediatrico Bambino Gesù (1702_OPBG_2018), Rome. Physical assessments were performed by experienced clinical geneticists and neurologists. Clinical data, pictures, DNA specimens, and other biological material were collected, used and stored after signed informed consents from the participating subjects/families were obtained. Permission to publish clinical pictures was obtained from 3 out of the 5 subjects (Pt2, Pt4, and Pt5).
2.2 Genomic analyses
Whole exome sequencing (WES) was performed in subjects Pt1, Pt2, and Pt3 by using capture kits and platforms specified in Table S1. Sequencing data were analyzed using pipelines based on the GATK Best Practices,12 and output is provided in Table S1. In brief, the UCSC GRCh37/hg19 version of genome assembly was used as a reference for reads alignment by means of BWA-MEM13 (https://doi.org/10.48550/arXiv.1303.3997) tool and subsequent variant calling. High-quality variants were prioritized against public databases (dbSNP150 and gnomAD V.2.0.1) retaining variants with unknown frequency or MAF < 0.1%. SnpEff v.4.314 and dbNSFP v.3.515 were used for variant functional annotations alongside combined annotation-dependent depletion (CADD) v.1.4,16 Mendelian Clinically Applicable Pathogenicity (M-CAP) v.1.017 and Intervar v.2.0.118 for in silico functional impact predictions. Graphic representation of the tolerance scores of affected amino acids was obtained by MetaDome.19 Clinically relevant single nucleotide variants (SNV) were also classified by referring to the American College of Medical Genetics and Genomics (ACMG) recommendations for variant classification.20 For sequencing statistics and metrics see Table S1.
Array-based comparative genomic hybridization analyses were performed in all subjects with a resolution ranging from ~30 to 130 kb. Genome-wide single-nucleotide polymorphism (SNP) array analysis was performed using the Human-Omni-Express Bead Chip array (Illumina, Inc., San Diego, CA, USA) which contains over 700 000 markers (mean resolution 30 kb), according to the manufacturer's instructions. In brief, 200 ng of genomic DNA was amplified, fractionated, hybridized, and fluorescently tagged. After scanning of the slides, further analysis was carried out using the software (GenomeStudio 2.0) provided by Illumina. Genotype and copy number were calculated by determination of B-allele frequency and log2 R ratio. Genomic coordinates are based on GRCh37/hg19 (Table S2). Array-based Comparative Genomic Hybridization (array-CGH) analysis was performed using the SurePrint G3 Human CGH Microarray kit 8x60K (Agilent, Santa Clara, CA, USA) with an average resolution of 130 Kb, following the manufacturer's protocol. Nucleotide designations were assigned according to the hg19/GRCh37 assembly of the human genome. For statistics and metrics, see Table S2.
Structural modeling of the NOT-box domain of the protein was performed using the crystal structure of NOT module of human CCr-not complex (PDB 4C0D, chain-B).21 Graphical representation was generated by using Pymol packaged software (www.pymol.org/pymol).
Statistical analysis and Fisher's exact test were conducted by using JavaStat_2-way Contingency Table Analysis (https://statpages.info/ctab2x2.html).
3 RESULTS
3.1 Molecular assessments
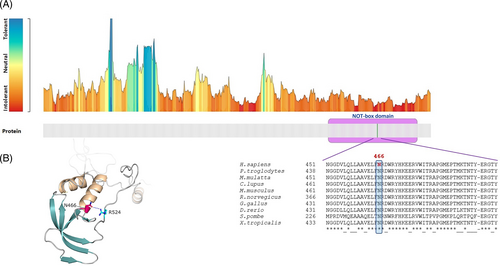
Five de novo molecular events affecting CNOT2 (HGNC:7878) were identified in five unrelated individuals with an undiagnosed neurodevelopment condition (Table 1). Subject 1 was enrolled at the Ospedale Pediatrico Bambino Gesù “Undiagnosed Patients Program,” an initiative directed to accelerate diagnosis for patients with unsolved clinical conditions. Additional subjects with an overlapping phenotype were identified by using online matchmaking empowered by the GeneMatcher platform22 and networking. Clinical data and molecular details are provided in Tables 1, 2, S5 and Supporting Information. Subject 1 carried a truncating variant (c.1482T>G, p.Tyr494Ter), Subject 2 showed a stop-loss variant predicting a 64 residue-extension of the encoded protein (c.1622A>T, p.Ter541Leuext*65). Both variants were not annotated in public databases (gnomAD_v2.0.1; ExaC_v0.3.1), and were classified as likely pathogenic according to the ACMG recommendations. While the former variant (c.1482T>G, p.Tyr494Ter) was predicted to generate a premature termination of the protein (CADD phred score: 36; ACMG criteria: PVS1, PM2), the latter (c.1622A>T, p.Ter541Leuext*65) was predicted to cause the loss of stop codon and extension of the coding sequence for additional 64 codons at 3′-UTR before termination (CADD phred score: 13.2; ACMG criteria: PM4, PM2, BP4). A missense change (chr12:70739964A>C, GRCh37.p13, c.1396A>C, p.Asn466His) was identified in an individual (Pt3), and segregation analysis confirmed its de novo origin. The amino acid substitution p.Asn466His had not previously been reported in public databases and was predicted to exert a damaging effect on the mature protein (CADD phred score: 24.1; M-CAP score: 0.026). According to the ACMG criteria, the change was classified as variant of uncertain significance (VUS) (PM2, PP3), however, the assessment of the 3D model of the protein indicated that it is able to considerably destabilize the structure of NOT-box domain (residues 437–540) (Figure 4a,b), compromising the protein function. The highly conserved Asn466 residue was predicted to be intolerant to changes and its substitution with histidine was expected to cause the loss of a H-bond with Arg524 within the domain (Figure 4b), resulting in perturbation of intradomain interactions, destabilization of the CNOT2 structure, with subsequent loss of protein function.
| Genomic coordinates (GRCh37) | Variant type | Coding variant (NM_014515.7) | Exon(s) | Amino acid change | Inheritance | rsID | CADD (phred_score) | ACMG (class) | Predicted effect on CNOT2 protein |
|---|---|---|---|---|---|---|---|---|---|
| chr12:70739964A>C | SNV | c.1396A>C | 15 | p.Asn466His | De novo | – | 24.1 | 3 | Loss-of-function (predicted) |
| chr12:70740050 T>G | SNV | c.1482 T>G | 15 | p.Tyr494Ter | De novo | – | 36.0 | 5 | Premature termination |
| chr12:70747694A>T | SNV | c.1622A>T | 16 | p.Ter541Leuext*65 | De novo | – | 13.2 | 5 | Stop loss and extension of the protein up to 65 codons at 3′UTR |
| chr12:70647181-77 916 092 | SV—7.27 Mb deletion | – | Entire gene | – | De novo | – | – | 4 | Haploinsufficiency |
| chr12:70733662-70 757 341 | SV—Intragenic deletion | – | 12–16 | – | De novo | – | – | 5 | Haploinsufficiency |
- Note: ACMG classification: 1, Benign; 2, Likely benign; 3, Uncertain significance; 4, Likely pathogenic; 5, Pathogenic.
| Pt1 | Pt2 | Pt3 | Pt4 | Pt5 | CNOT2 variant/Intragenic deletion | 12q15 Structural variation | Total N (%) | |
|---|---|---|---|---|---|---|---|---|
| Reference(s) | This study | This study | This study | This study | This study | 9-11 | 1-5 | |
| CNOT2 variant/structural variant | p.Tyr494Ter | p.Ter541Leuext*65 | p.Asn466His | Intragenic deletion | 7.27 Mb deletion | p.Lys316Ter Int3-3′UTR del 5′UTR-Int3 del |
12q15 deletion (0.74–10.21 Mb) | |
| Inheritance | De novo | De novo | De novo | De novo | De novo | De novo | De novo | |
| Age at last examination | 10 months | 12 years | 20 years | 3 years | 6 years | 6–40 years | 5–29 years | |
| Gender | M | F | F | M | M | 3 M | 5 F; 2 M | |
| Ethnicity | Saudi Arabia/Syria | Reunion Island | Saudi Arabia | Han Chinese/Caucasian | Caucasian | Japanese/Caucasian | Caucasian | |
| Prenatal growth | ||||||||
| Gestational week | 36 | 40 | At term | 35 + 4 | 34 C-s | At term | At term | |
| Growth retardation (< −2 SD) | No | No | Yes | No | No | No | 1/7 | 2/15 (13) |
| OFC restriction (< −2 SD) | No | No | No | No | N/A | No | No | (0) |
| Postnatal growth | ||||||||
| Growth retardation (<2 SD) | Yes | No | No | N/A | No | No | 1/4 | 2/11 (18) |
| OFC restriction (<2 SD) | No | No | No | No | Yes | No | No | 1/15 (8) |
| Neurological findings | ||||||||
| DD/ID | Yes | Yes | Yes | Yes | Yes | 3/3 | 7/7 | 15/15 (100) |
| Speech delay | Yes | Yes | No | Yes | Yes | 3/3 | 6/7 | 13/15 (87) |
| Walking delay | Yes | Yes | Yes | Yes | Yes | 3/3 | 5/7 | 13/15 (87) |
| Hypotonia | No | Yes | Yes, MW | Yes | Yes | 1/1 | 4/6 | 9/12 (75) |
| Nasal speech | N/A | Yes | N/A | N/A | No | 1/3 | 6/7 | 8/12 (67) |
| Feeding problems | No | No | No | No | No | 1/3 | 5/7 | 6/15 (40) |
| Behavioral abnormalities | N/A | Yes, HA | No | No | Yes, AD, SD | 1/1 | 1/1 | 4/6 (67) |
| MRI/CT changes | No | No | No | No | Yes, DCC, RWM, TB, HCV | No, (1 subject, IBV) | No | 1/15 (7) |
| Seizures/Epilepsy/EEG abnormalities | No | No | No | No | No | 1/3, SE | N/A | 1/8 (12) |
| Craniofacial features | ||||||||
| High forehead | Yes | Yes | N/A | Yes | Yes | 2/3 | 7/7 | 13/14 (93) |
| Flat forehead | No | Yes | N/A | Yes | Yes | 2/3 | 4/5 | 9/12 (75) |
| Straight eyebrows | No | Yes | No | No | No | 3/3 | 5/7 | 9/15 (60) |
| Thick/bushy eyebrows | No | Yes | No | Yes | No | 3/3 | 2/5 | 7/13 (54) |
| Deep set eyes | Yes | Yes | Yes | Yes | Yes | 2/2 | 2/5 | 9/12 (75) |
| Upslanting palpebral fissures | Yes | Yes | No | Yes | No | 3/3 | 3/5 | 9/13 (69) |
| Long eyelashes | Yes | No | Yes | Yes | Yes | 2/3 | 3/5 | 9/13 (69) |
| Flat face/midface hypoplasia | Yes | No | N/A | Yes | No | 1/3 | 4/7 | 7/14 (50) |
| Short triangular nose | Yes | Yes | N/A | Yes | Yes | 3/3 | 5/5 | 12/12 (100) |
| Anteverted nares | Yes | Yes | N/A | Yes | Yes | 3/3 | 5/5 | 12/12 (100) |
| Long philtrum | No | No | No | No | No | 1/3 | 4/7 | 5/15 (33) |
| Thin upper lip | No | No | N/A | Yes | Yes | 3/3 | 3/5 | 8/12 (67) |
| Micrognathia/retrognathia | Yes | Yes | N/A | Yes | No | 3/3 | 4/7 | 10/14 (71) |
| High arched palate | N/A | Yes | N/A | N/A | N/A | 1/2 | 1/2 | 3/5 (60) |
| Low set ears | Yes | Yes | Yes | Yes | No | 1/3 | 4/7 | 9/15 (60) |
| Hand/foot anomalies | ||||||||
| Brachydactyly | No | No | No | Yes | Yes | 2/3 | 1/6 | 5/14 (36) |
| Slender fingers | Yes | Yes | No | No | No | 1/3 | 1/6 | 4/14 (29) |
| Clino/campto-dactyly | No | No | No | Yes | No | 1/3 | 3/5 | 5/13 (38) |
| Skeletal defects | ||||||||
| Scoliosis/kyphosis | No | Yes | Yes | Yes | Yes | 1/3 | 2/7 | 7/15 (47) |
| Vertebral/sacral defects | No | No | Yes | No | No | 1/2 | 2/7 | 4/15 (27) |
| Left/Right asymmetry | No | No | No | No | No | 1/3 | 2/7 | 3/15 (20) |
| Joint hypermobility | No | Yes | No | No | No | No | No | 1/15 (7) |
| Hyperextensible skin | No | No | No | N/A | No | No | 1/1 | 1/8 (12) |
| Cardiac anomalies | No | No | No | No | Yes, PS, DV | 2/3, PS, DV, MI | 1/7, VSD | 4/15 (27) |
| Genitourinary anomalies | No | No | No | Yes, PN | No | 1/3, RD | No | 2/15 (13) |
| Additional anomalies | LM, LL, CS | Bf, E, My, ACKL | SN, St, LM | LM (3/3) |
- Abbreviations: ACKL, Augmented creatine kinase levels (serum); AD, attention deficit; Bf, blepharophimosis; C-s, C-section delivery; CS, cup-shaped ears; DCC, dysmorphic corpus callosum; DV, dysplastic valve; E, exotropia; HA, hetero-aggressiveness; HCV, hypoplasia cerebellar vermis; IBV, irregular border of ventricles; LL, large ear-lobe; LM, large mouth; MI, mitral insufficiency; MW, muscle weakness; My, myotonia; N/A, not assessed; No, feature absent; PN, pyelonephritis; PS, pulmonic stenosis; RD, renal dysplasia; RWM, reduced white matter; SD, sleep disturbances; SE, seizures, SLE, sparse lateral eyebrows; SN, supernumerary nipples; St, strabismus; TB, thin brainstem; TUL, thin upper-lip/absent Cupid's bow; US, upslanted palpebral fissures; VSD, ventricular septal defect; Yes, feature present.
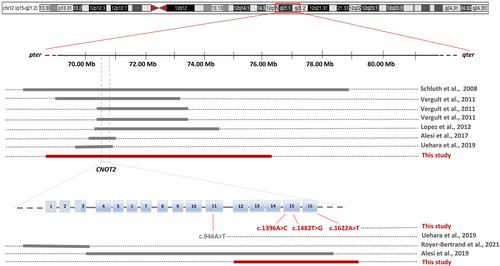
Chromosome and array-based analyses were carried out for all subjects. While the occurrence of clinically relevant structural variations (SVs) were ruled out in three subjects (Pt1–Pt3), microdeletions at 12q15 locus were identified in two individuals (Pt4 and Pt5) (Table 1). Subject 4 (Pt4) carried a 23.7 kb intragenic microdeletion, arr[GRCh37] 12q15(70 733 662_70 757 341)x1 dn, which was predicted to cause loss of five exons of the gene from 12 to 16. The SV had not previously been reported in public databases and was classified as likely pathogenic (http://dgv.tcag.ca/dgv/app/home). Subject 5 (Pt5) showed a de novo large 7.27 Mbp deletion (12q15q21.2), arr[GRCh37] 12q15(70 647 181_77 916 092)x1 dn, encompassing CNOT2 and other 25 genes (Table S2), of which, two disease-causing genes (BBS10 and TPH2) (Table S3).
3.2 Clinical profile
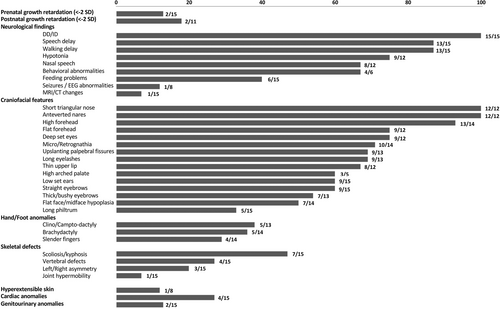
Subjects carrying heterozygous variants within the CNOT2 coding sequence (Pt1–Pt4) showed an overlapping phenotype, which was also congruent with the clinical profile of the individual with 7.27 Mbp 12q15 deletion (Pt5) (Figure 1). To verify whether IDNADFS and chromosome 12q15 deletion syndrome could be considered a unique disorder, the major clinical features of patients from the present and published series (n = 15, 8 males and 7 females) were compared (Table 2, Figure S1). We first clustered patients in two classes (intragenic CNOT2 variants and large structural deletions encompassing the gene) (Figure S1). Interestingly, when comparing 21 suggestive clinical items, none showed significant difference (Fishers' test analyses) between the two classes (Figure S1), confirming that CNOT2 haploinsufficiency underlies both IDNADFS and chromosome 12q15 deletion syndrome.
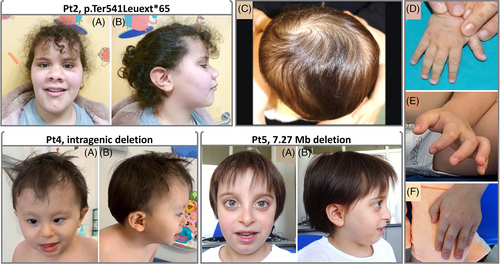
Based on these results, we systematically assessed the features of all affected individuals (n = 15) to demarcate the clinical profile of the CNOT2-related disorder. Clinical data and frequency of features are provided in Tables 2, S5, Figure 2 and Supporting Information. Prenatal and postnatal growth retardation (<−2 SD) were observed in four subjects (Tables 2 and S5). Neurologic features included DD/ID (15/15), speech delay (13/15), hypotonia (9/12), delay in walking (13/15), and nasal speech (8/12). Feeding problems were documented in some patients (6/15). Brain anatomic changes (i.e., reduction of white matter and hypoplasia of the corpus callosum and cerebellum) were documented in single cases (1/15). Behavioral anomalies were also sporadically seen in the present cohort (4/6), even though, this item was not specifically investigated in all subjects. Electroencephalographic (EEG) abnormalities were found only in a subject (1/8). Facial features were recurrent and recognizable among the affected subjects regardless their intragenic or genomic CNOT2 variation. While microcephaly (with occipito-frontal circumference <−2 SD) was rarely observed, facial dysmorphisms, including high and flat forehead (13/14 and 9/12, respectively), straight and thick eyebrows (9/15 and 7/13, respectively), deep set eyes (9/12), upslanted palpebral fissures (9/13), long eyelashes (9/13), flat face/midface hypoplasia (7/14), short triangular nose and anteverted nares (12/12), long philtrum (5/15), thin upper lip (8/12), and micro/retro-gnathia (10/14) were recognized in most individuals. Low-set, mainly posteriorly rotated ears were also found in half of the subjects (9/15). Hand/feet abnormalities included either brachydactyly (5/14), or slender fingers (4/14), and clino/campto-dactyly (5/13) (Figure 1, Tables 2 and S5). Variable skeletal defects were also documented, including scoliosis/kyphosis (7/15), and nonspecific vertebral/sacral defects (4/15). Of note, abnormal skull shape (brachycephaly, oxycephaly, and plagiocephaly) (Figure 1), left/right asymmetry of pelvis and lower limbs (3/15), were noticed in some individuals. Joint hyperlaxity was reported in one patient. Other anatomical defects, including congenital heart defects (CHD) (ventricular septum defect, supra-valvular and valvular pulmonic stenosis, and dysplastic valves, 4/15), and genitourinary/kidney anomalies (renal dysplasia and early onset pyelonephritis, 2/15), and hyperextensible skin (1/8) were also documented in a few individuals (Tables 2 and S5).
4 DISCUSSION
IDNADFS is a neurodevelopmental disorder characterized by DD/ID, hypotonia, variable skeletal defects, and craniofacial dysmorphism and is caused by CNOT2 heterozygous variants but also related to chromosome 12q15 deletion syndrome.4, 9, 10 However, given the small number of reported individuals, this association is still debated.5 Here, we expanded the series describing five additional individuals with heterozygous SNVs and structural rearrangements affecting CNOT2 (Table 1). These subjects showed an overlapping phenotype (Figure 1 and Table 2), which was also consistent with the clinical features of 10 reported individuals with IDNADFS or chromosome 12q15 deletion syndrome.1-6, 10, 11 In order to verify the role of CNOT2 as the critical gene of chromosome 12q15 deletion syndrome, we compared the clinical features of 15 individuals with CNOT2 haploinsufficiency from present and published series. Fifteen molecular events affecting CNOT2, including two non-sense (p.Tyr494Ter, p.Lys316Ter) variants, one stop-loss variant (p.Ter541Leuext*65), a single missense change (p.Asn466His), three disruptive intragenic deletions affecting either the 5′- or 3′-end, and eight deletions within 12q15 encompassing CNOT2, were considered for the phenotype comparison (Tables 1 and 2, Figure 3). Fishers' exact test performed on 14 facial features and 7 additional relevant clinical signs (i.e., DD/ID, speech delay, nasal speech, scoliosis, CHD, and feeding problems) (Figure S1) indicated that loss of CNOT2 is the major contributing event of chromosome 12q15 deletion syndrome.
The entire clinical profile of IDNADFS is not fully delineated yet, and the occurrence of a subset of findings (i.e., pectus excavatum, delayed bone age, chronic renal failure, and CHDs) is still debated.5, 10 Given the evidence of a unique nosological entity caused by CNOT2 haploinsufficiency, we reviewed 41 features characterizing the clinical spectrum of CNOT2-related disorder to clinically delineate this condition (Tables 2 and S5). The most consistent features are ID (ranging from mild to severe), delay in reaching language and motor skills milestones, hypotonia, and a distinctive craniofacial appearance. These findings can be considered as portraying the disorder. Nasal speech has been frequently noticed in the affected subjects (67%) and can be also considered characteristic. In the presence of ID, specific brain structural abnormalities have not been documented in the present series, suggesting that the learning disability is possibly due to impaired intellectual functioning. Of note, behavioral/psychiatric problems have also been observed in the disorder even though these issues have not been fully investigated. Feeding problems may also occur (40%) resulting in failure to thrive. Seizures or EEG abnormalities have been sporadically reported11 and might be the result of an incidental finding. Craniofacial dysmorphisms are recurrent among subjects and quite distinctive (Figure 1). Facial features include high and flat forehead (resulting in an abnormal skull shape in some subjects), straight and bushy eyebrows, deep-set eyes, upslanted palpebral fissures, short and triangular nose with anteverted nares, thin upper lip, mild micro/retro-gnathia, and low-set ears (Figure 1, Tables 2 and S5). Brachydactyly, slender fingers, clino/campto-dactyly, and broad thumbs are reported in a minority of individuals. Skeletal anomalies are variable and include scoliosis/kyphosis and vertebral/sacral defects in 47% and 27% of subjects, respectively. Notably, abnormal shape of the cranium and left/right asymmetry have sporadically been evidenced in the present cohort (Figures 1, Tables 2 and S5). Other less common features include CHD and genitourinary anomalies, and hyperextensible skin (Figure 2 and Table 2). A more extended series of cases, however, is required to corroborate their occurrence and relevance in the disorder. Overall, data from the present study allow us to confirm that the core phenotype of CNOT2-related disorder is characterized by DD/ID, speech delay with nasal speech, hypotonia and a distinctive craniofacial appearance.
CNOT2 is a component of the CCR4-NOT complex, which is an evolutionary conserved multisubunit protein complex involved in various cellular processes, including cell growth, DNA repair, mRNA export, deadenylation and decay, histone methylation, and protein quality control.23, 24 The complex contains up to 11 different subunits (CNOT1-11 and CNOT6L), each with a specific function.25 For example, while CNOT6, CNOT6L, CNOT7, and CNOT8 subunits are required for the deadenylase activity,26 CNOT10-CNOT11 module and CNOT4 are essential for recruitment of target mRNAs.27, 28 On the other hand, CNOT1, CNOT2, CNOT3, and CNOT9 do not possess catalytic activities but exert a control function of the entire complex by mediating key intermolecular interactions between subunits, and have an overall scaffold role.29, 30 Of note, CNOT2 and CNOT3 are structurally similar displaying a unique conserved domain, the “NOT-box” located at the C-terminus, which is crucial for their reciprocal interaction.21 Depletion of CNOT2 in HeLa cells extensively decreases the expression of CNOT3 and other CNOTs members, and impairs the integrity and function of the entire CCR4-NOT complex.29 Consistently, dominant acting mutations disrupting other members of the complex (such as CNOT1 and CNOT3) cause neurodevelopment syndromes (OMIM#619033 and OMIM#618672, respectively), which interestingly share several clinical features with IDNADFS (i.e., DD/ID, hypotonia, speech delay, behavioral anomalies, and variable skeletal abnormalities)31, 32 (Table S4). Noteworthy, some craniofacial dysmorphisms (i.e., prominent/high forehead, upslanted palpebral fissures, anteverted nares, long philtrum, thin upper lip, low-set ears), and brachydactyly are also characteristics of the CNOT3-related disorder (IDDSADF, OMIM#618672) (Table S4). These data suggest the existence of a phenotypic continuum caused by defective function of the CCR4-NOT complex, for which an accurate differential diagnosis should be considered.
In the present study, we have reported five additional de novo events affecting CNOT2. In addition to the large microdeletions at the 12q15 locus, the novel intragenic deletion and the truncating variants further confirm that CNOT2 mutations likely result in haploinsufficiency. Of note, the identified 7.27 Mbp deletion encompasses CNOT2 and other 25 genes (Pt5, Table S3), none of which supports a causative role in the disorder (Table S3). Finally, we provide evidence that IDNADFS may result from loss-of-function missense variants affecting the gene. In particular, the identified change (p.Asn466His) is believed to dramatically perturb the CNOT2 structure and function, resulting in a disease-causing mutation (Figure 4). In vitro or in vivo functional validations of this variant are required to confirm this assumption.
In conclusion, our data demonstrate that IDNADFS strictly overlaps with chromosome 12q15 deletion syndrome, confirming that CNOT2 haploinsufficiency and/or its loss of function are the major events underlying a unique disorder. The clinical profile of the CNOT2-related disorder is mainly characterized by DD/ID, speech delay with nasal speech, hypotonia and a distinctive facial appearance, and shares a subset of features with other CNOTs-related disorders, outlining a new family of neurodevelopmental disorders.
AUTHOR CONTRIBUTIONS
Conceptualization: Marcello Niceta. Data curation (genetic and clinical investigations): Simone Pizzi, Francesca Inzana, Angela Peron, Somayeh Bakhtiari, Mathilde Nizon, Jonathan Levy, Alejandro Ferrer, Cecilia Mancini, Benjamin Cogné, Eric W. Klee, Pavel Pichurin, Francesca Clementina Radio, Emanuele Agolini, Dario Cocciadiferro, Antonio Novelli, Mustafa A. Salih, Maria Paola Recalcati, Rosangela Arancio, Marianne Besnard, Anne-Claude Tabet, Michael C. Kruer, and Marcello Niceta. Project administration: Bruno Dallapiccola, Marco Tartaglia. Preparation of the original draft: Marcello Niceta, Simone Pizzi. Reviewing and commenting the manuscript: All authors. Fine tuning and final editing of the manuscript: Marcello Niceta, Manuela Priolo, Bruno Dallapiccola, Marco Tartaglia.
ACKNOWLEDGMENTS
All authors are thankful to Dr. Camilla Capelletti (Bambino Gesù Children's Hospital, IRCCS, Rome, Italy) and Dr. Muddathir H. Hamad (King Saud University, Riyadh, Saudi Arabi) for their contribution in collecting data, and thankful to probands and their parents for the kind availability. Open access funding provided by BIBLIOSAN.
FUNDING INFORMATION
This work was supported, in part, by grants from the Italian Ministry of Health (Ricerca 5 per Mille, to M.T.), and Fondazione Bambino Gesù (Vite Coraggiose).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




