Application of whole-exome sequencing for detecting copy number variants in CMT1A/HNPP
Abstract
Large insertions and deletions (indels), including copy number variations (CNVs), are commonly seen in many diseases. Standard approaches for indel detection rely on well-established methods such as qPCR or short tandem repeat (STR) markers. Recently, a number of tools for CNV detection based on next-generation sequencing (NGS) data have also been developed; however, use of these methods is limited. Here, we used whole-exome sequencing (WES) in patients previously diagnosed with CMT1A or HNPP using STR markers to evaluate the ability of WES to improve the clinical diagnosis. Patients were evaluated utilizing three CNV detection tools including CONIFER, ExomeCNV and CEQer, and array comparative genomic hybridization (aCGH). We identified a breakpoint region at 17p11.2-p12 in patients with CMT1A and HNPP. CNV detection levels were similar in both 6 Gb (mean read depth = 80×) and 17 Gb (mean read depth = 190×) data. Taken together, these data suggest that 6 Gb WES data are sufficient to reveal the genetic causes of various diseases and can be used to estimate single mutations, indels, and CNVs simultaneously. Furthermore, our data strongly indicate that CNV detection by NGS is a rapid and cost-effective method for clinical diagnosis of genetically heterogeneous disorders such as CMT neuropathy.
Structural variants including copy number variation (CNV) and insertions and deletions (indel) have been highlighted as the causes of genetic disorders. Recently, it has been reported that CNVs significantly contribute to various diseases such as neurodevelopmental disorders, intellectual disabilities and numerous cancers 1-3. In particular, Charcot–Marie–Tooth disease type 1A (CMT1A; MIM 118220) and hereditary neuropathy with liability to pressure palsies (HNPP; MIM 162500) are caused by duplication and deletion, respectively, of 1.4 Mb region including the peripheral myelin protein 22 gene (PMP22; MIM 601097) on 17p11.2-p12, resulting from unequal crossover during meiosis 4.
Since the advent of next-generation sequencing (NGS)-based technologies in 2008 5, the ability to perform comprehensive genomic analyses has accelerated dramatically, allowing for accurate characterization of genetic diseases at increasingly low costs. When coupled with advances in genomic capture techniques, whole-exome sequencing (WES) has become an attractive alternative for variant detection with both high specificity and sensitivity 6. Although whole genome sequencing (WGS) is used primarily to detect large indels, including CNVs or loss of heterozygosity (LOH), numerous algorithms applicable to WES data allow estimation of structural variations 7.
In this study, we showed the feasibility of WES for detecting the underlying genetic causes in not only difficult-to-diagnose patients, but also various types of heterogeneous disorders at once. As the CNV regions in our samples consisted of large indels >5 kb previously validated using STR markers, we applied three WES-based approaches for CNV detection, ExomeCNV, CONIFER, and CEQer, and compared these approaches with aCGH, the current gold standard for CNV detection. Using these approaches, we were able to accurately identify the chromosomal breakpoint within the 17p11.2–p12 region in CMT1A/HNPP patients. Lastly, we compared the outcomes of WES-based approaches at mean read depths of 6 vs 17 Gb data to find out if the generally used read depth (6 Gb) is enough for accurate CNV estimation.
Patients and methods
Subjects
This study examined three patients (FC383, FC388, and HN129); two of them were affected by CMT1A, and the other was affected by HNPP (HN129). The clinical evaluation of these patients was performed by two independent neurologists. Written informed consent was obtained from all participants, including the three controls, according to the protocols approved by the Institutional Review Board of Ewha Woman's University, Mokdong Hospital, and the Korea National Institutes of Health (KNIH).
Genetic analysis
NGS-based tools were used to analyze three patients who had been diagnosed previously with CMT1A and HNPP using six microsatellite markers 8. The genetic causes of CMT1A/HNPP in each of these three patients (FC383, FC388, and HN129) were full duplication, partial duplication, and deletion, respectively, mapping to 17p11.2–p12, including the entire PMP22 gene.
Whole-exome sequencing
We performed targeted capture and massively parallel sequencing for all three individuals. Whole exomes were captured using the SeqCap EZ Human Exome Library v2.0 (Roche/NimbleGen, Madison, WI) to the 6 Gb data and the Agilent SureSelect XT V4 to the 17 Gb data (Table S1 and S2, Supporting information). Captured libraries were sequenced using the Illumina HiSeq 2000 system (Illumina, San Diego, CA) according to the manufacturer's protocols. Reads were mapped to the reference human genome (GRCh37, UCSC hg19) using the Burrows-Wheeler Aligner (http://bio-bwa.Sourceforge.net/).
Whole-exome CNV analysis
WES data were analyzed using three individual CNV calling algorithms based on read depth: (i) ExomeCNV 9, (ii) CONIFER 10, and (iii) CEQer 11. On CONIFER, a pooled sample calling approach was used as input with three controls (FC283-5, FC417-2, and FC378-3) for 6 Gb dataset and two controls (FC283-5 and FC417-2) for the 17 Gb dataset. For ExomeCNV and CEQer, a case–control sample calling approach, was used, along with a single control (FC283-5) in CEQer and two controls (FC283-5 and FC417-2) in ExomeCNV.
Oligonucleotide-based aCGH analysis
Four samples, including three cases and one control (FC283-5), were also analyzed by aCGH (Agilent SurePrintG3 2 × 400 k). Data analysis was performed on the Agilent Genomic Workbench 7.0 using the ADM-2 algorithm with a default threshold of 6.
Results
CNV detection by WES
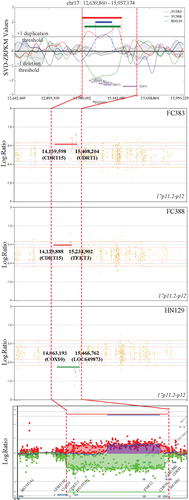
To determine accurate breakpoints for duplication and deletion events in CMT1A/HNPP patients by WES, a 1.4 Mb region, which is delimited by two 24 kb low copy number repeats (CMT1A-REPs) on 17p11.2-p12, was targeted for downstream analysis. Analyses of CNV within this region were performed using three individual WES-based CNV algorithms (Fig. 1). The three CNV detection tools used here rely on a read-depth approach that determines the mapping ratio of read counts relative to a reference genome. Breakpoints of duplication and deletion events for CMT1A and HNPP, as determined using these methods, are shown in Table 1.
| aCGH-WES | ||||||
|---|---|---|---|---|---|---|
| Start | End | Start | End | |||
| aCGH | FC383 | 14,093,244 | 15,479,524 | |||
| FC388 | 14,649,346 | 15,366,750 | ||||
| HN129 | 14,086,954 | 15,442,069 | ||||
| CONIFER | FC383 | 6 Gb | 14,005,386 | 15,466,820 | 87,858 | 12,704 |
| 17 Gb | 14,063,167 | 15,443,972 | 30,077 | 35,552 | ||
| FC388 | 6 Gb | 14,683,140 | 15,231,420 | −33,794 | 135,330 | |
| 17 Gb | 14,683,115 | 15,341,585 | −33,769 | 25,165 | ||
| HN129 | 6 Gb | 14,110,101 | 15,457,174 | −23,147 | −15,105 | |
| 17 Gb | 14,063,167 | 15,449,175 | 23,787 | −7,106 | ||
| CEQer | FC383 | 6 Gb | 14,139,598 | 15,498,204 | −46,354 | −18,680 |
| 17 Gb | 14,095,305 | 15,449,230 | −2,061 | 30,294 | ||
| FC388 | 6 Gb | 14,139,888 | 15,234,902 | 509,458 | 131,848 | |
| 17 Gb | 14,063,193 | 15,234,323 | 586,153 | 132,427 | ||
| HN129 | 6 Gb | 14,063,193 | 15,466,762 | 23,761 | −24,693 | |
| 17 Gb | 14,095,305 | 15,449,230 | −8,351 | −7,161 | ||
| ExomeCNV | FC383 | 6 Gb | 14,095,266 | 15,457,018 | −2,022 | 22,506 |
| 17 Gb | 14,095,219 | 15,492,578 | −1,975 | −13,054 | ||
| FC388 | 6 Gb | 14,673,441 | 15,343,623 | −24,095 | 23,127 | |
| 17 Gb | 14,673,381 | 15,468,878 | −24,035 | −102,128 | ||
| HN129 | 6 Gb | 14,095,266 | 15,492,578 | −8,312 | −50,509 | |
| 17 Gb | 14,095,219 | 15,468,878 | −8,265 | −26,809 | ||
Performance of WES methods relative to the aCGH
We performed aCGH, as a gold standard, on four samples and compared its effectiveness with those of our three WES-based CNV estimation algorithms. All three CNV detection tools exhibited high correlation relative to aCGH (Fig. 1).
Next, we compared the resolution of CNV breakpoints within the CMT1A REP region using WES and aCGH-based platforms. In the high-resolution microarray, the duplication or deletion of target regions was detected within 17p11.2–p12 for both CMT1A and HNPP (Fig. 2). Similar results were obtained using WES-based methods, with <1% difference in breakpoint locations between the methods, relative to the full-length chromosome 17 (81,195,210 bp). The largest difference between WES and aCGH breakpoints was seen for case FC388, who harbored a partial duplication, while the smallest difference was seen for case HN129, for both the 6 and 17 Gb datasets (Fig. 2a). In terms of analysis methods, the CEQer exhibited the greatest difference and ExomeCNV the least relative to aCGH (Fig. 2b). Of the three CNV detection tools used in this study, ExomeCNV was the most effective at replicating the aCGH results.
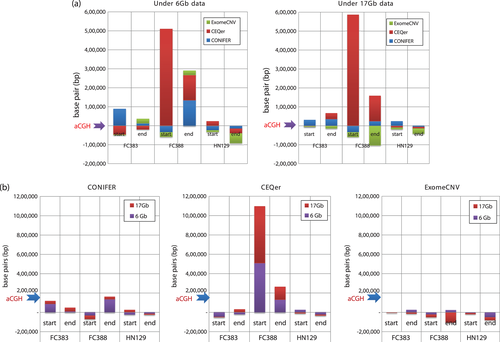
Comparisons based on differences in mean read depth of total yield
We examined the effectiveness of WES-based methods relative to mean read depth of total yield (6 and 17 Gb data) to determine the importance of read depth for CNV applications. Moreover, this analysis helped establish baseline criteria for WES-based analyses, which currently rely upon 60–80× mean read depth for most applications. Exome capture platforms were shown to perform well at both the 6 and 17 Gb levels. While differences were detected between the SeqCap EZ human exome library v2.0 and Agilent SureSelectXT V4 kit, these differences were not systematically significant.
Discussion
Here, we evaluated the feasibility of WES-based methods for identifying large insertions and deletions in CMT1A/HNPP patients. We compared the results of three individual CNV estimation algorithms with those of an aCGH platform, which is considered the gold standard for high-throughput CNV detection. This analysis revealed a high degree of reproducibility between the methods, confirming the effectiveness of WES-based platforms as diagnostic tests for CNV-caused diseases.
The three read depth-based CNV detection tools used here were selected based on previous reports 12, 13. All showed strong reproducibility relative to aCGH and high detection of CNVs within the CMT1A-REP region, revealing ExomeCNV as the effective method relative to aCGH in our study. This suggests that ExomeCNV is a suitable option for detecting germline variations, despite being designed for detecting CNVs on cancer. Likewise, CONIFER, capable of identifying rare genetic variants particularly within large exome datasets, compared well with aCGH. It may be because this tool adjusts for positional fluctuations associated with targeted capture sequencing by applying a Z-score. CEQer, graphical program for CNV detection at the whole-exome level, was less capable of reproducing the aCGH results; yet, it has the most user-friendly interface of all three methods, as it can be run using a standard Windows-based operating system, as well as accepting BAM/pileup formats of the sequencing datasets.
For many applications, WES is carried out at a mean read depth of 80×. We wanted to see whether higher read depth would result in higher resolution of CNV events, with better definition of the breakpoint sites. Our data indicated that the differences in mean read depth of total yield did not affect resolution or our ability to detect genomic variations.
For WES-CNV analysis, the validation of the CNVs is necessary due to high GC content, mapping artifacts, and algorithm-specific biases that can result in a high false positive rate, low sensitivity, and duplication and deletion biases. There are two typical methods with which to validate WES-based CNVs: (i) aCGH, an array-based platform, and (ii) qPCR or ddPCR at the molecular level 14.
Since the region of interest within our samples was targeted, we were able to test a variety of threshold values for both the CONIFER and CEQer methods. For CONIFER and CEQer, the default threshold was slightly adjusted. In CONIFER, we lowered the default threshold from ±1.5 to ±1.0 to better detect duplication and deletion events associated with CMT1A/HNPP. In CEQer, a lower cut-off value was required at the 6 Gb resolution in FC388. These data suggest that the detection accuracy of genomic variations can be improved by adjusting the thresholds of WES algorithms.
There are various genetic causes in CMT1A, including a point mutation in PMP22 identified in a Dutch cohort with CMT1A 15, as well as a heterozygous 186 kb duplication on 17p12 but outside of the PMP22 coding region 16. Although quantitative PCR remains the most common method for detecting both duplication and deletion events in PMP22 associated with CMT1A and HNPP, respectively 17, it is limited in its ability to identify genetic causes, such as SNPs and indels. WES therefore represents a powerful alternative capable of simultaneous detection of SNPs, indels, and CNVs, allowing for improved diagnosis of heterogeneous disorders such as CMT1A.
The exact identification of CMT causative mutations is important for preimplantation genetic diagnosis (PGD), and may play an important role in the application of personalized therapy in the future 18. We performed NGS analyses to identify the genetic causes of CMT in Korean patients, and we verified that genetic screening was essential to diagnose less recognizable CMT phenotypes 19. Given recent improvements in both the cost and accessibility of WES, these methods may soon replace traditional single gene tests. Based upon the data presented here, we suggest the adoption of more comprehensive screening methods, such as NGS, as new standards in genetic testing for CMT1A.
While there are several limitations to WES in terms of CNV detection due to the unequal spacing of exons throughout the genome, these issues are easily overcome, enabling efficient detection of many genetic diseases, including both heterogenous and monogenic disorders, many of which are caused by mutations within the coding regions. Taken together, our study demonstrate that a range of genomic alternations can be evaluated using a single platform. We therefore propose WES as a potent alternative for the study and diagnosis of heterogenous disorders, such as peripheral neuropathy.




