Search for ReCQL4 mutations in 39 patients genotyped for suspected Rothmund–Thomson/Baller-Gerold syndromes
Abstract
Three overlapping conditions, namely Rothmund–Thomson (RTS), Baller-Gerold (BGS) and RAPADILINO syndromes, have been attributed to RECQL4 mutations. Differential diagnoses depend on the clinical presentation, but the numbers of known genes remain low, leading to the widespread prescription of RECQL4 sequencing. The aim of our study was therefore to determine the best clinical indicators for the presence of RECQL4 mutations in a series of 39 patients referred for RECQL4 molecular analysis and belonging to the RTS (27 cases) and BGS (12 cases) spectrum. One or two deleterious RECQL4 mutations were found in 10/27 patients referred for RTS diagnosis. Clinical and molecular reevaluation led to a different diagnosis in 7/17 negative cases, including Clericuzio-type poikiloderma with neutropenia, hereditary sclerosing poikiloderma, and craniosynostosis/anal anomalies/porokeratosis. No RECQL4 mutations were found in the BGS group without poikiloderma, confirming that RECQL4 sequencing was not indicated in this phenotype. One chromosomal abnormality and one TWIST mutation was found in this cohort. This study highlights the search for differential diagnoses before the prescription of RECQL4 sequencing in this clinically heterogeneous group. The combination of clinically defined subgroups and next-generation sequencing will hopefully bring to light new molecular bases of syndromes with poikiloderma, as well as BGS without poikiloderma.
Three autosomal recessive disorders have been associated with mutations in the RECQL4 gene: Rothmund–Thomson (RTS; MIM268400), Baller-Gerold (BGS; MIM218600) and RAPADILINO syndromes (MIM266280). RTS is the most prevalent phenotype in association with RECQL4 mutations, with more than 300 cases in the literature so far 1, 2. The main clinical hallmark is poikiloderma and the clinical diagnosis of RTS is currently based on the unique time of onset, and the appearance and spreading of poikiloderma. RTS diagnosis could also be discussed in cases of atypical rashes if they are associated with at least two of the following clinical signs: sparse scalp hair, eyelashes, and/or eyebrows; small size; gastrointestinal disorders; radial ray defects; radiographic bone abnormalities that include dysplasia, absent or malformed bones, osteopenia, abnormal trabeculation; dental abnormalities; dysplastic or poorly formed nails; hyperkeratosis particularly of the soles of the feet; cataracts; skin and bone cancers 2. Two clinical RTS subtypes have been defined. RTSI is characterized by poikiloderma and juvenile cataracts, and absence of RECQL4 mutations. RTSII is characterized by poikiloderma, congenital abnormalities including bone defects and an increased risk of cancer. Deleterious RECQL4 mutations are found in 2/3 of cases 1. Differential diagnoses for RTS mainly include disorders with poikiloderma [Clericuzio-type poikiloderma with neutropenia (CPN; MIM604173), Kindler syndrome (MIM173650), hereditary sclerosing poikiloderma (HSP; 173700), dyskeratosis congenita, xeroderma pigmentosum], as well as other rare genodermatoses with prominent telangiectasia but not true poikiloderma (Bloom, Werner, ataxia-telangiectasia, and Cockayne syndromes).
BGS is characterized by craniosynostosis (usually coronal), skeletal dysplasia with radial ray defects, and growth retardation 3. Differential diagnoses for BGS include disorders exhibiting radial ray hypoplasia as a major component and occasional craniosynostosis (Fanconi anaemia, foetal valproate syndrome, VACTERL, SALL4-related disorders, Holt–Oram syndrome, thrombocytopenia-absent radius syndrome) and syndromes with predominant craniosynostosis, in particular Saethre–Chotzen syndrome (SCS, MIM101400; 4 5). RECQL4 mutations have only been found in a subset of patients with BGS and poikiloderma 3.
Because only a few genes have been assigned to syndromic poikiloderma or BGS, the search for RECQL4 mutations is often prescribed, given the risks of recurrence and of malignancy. From the 5-year experience of a laboratory offering RECQL4 screening as a diagnostic test, we analysed a cohort of 39 patients referred for RECQL4 screening in order to determine the best indicators for RECQL4 sequencing, as well as possible diagnosis in negative patients.
Patients and methods
Patients
The 39 included patients had been referred to our laboratory for RECQL4 analysis from 2008 to 2013 because of poikiloderma compatible with RTS (cohort 1, 27/39 patients), or multiple congenital abnormalities with craniosynostosis and/or radial ray defects compatible with BGS without poikiloderma (cohort 2, 12/39 patients). All the patients provided written informed consent.
Methods
RECQL4, C16orf57/USB1 and TWIST1 sequencing analysis
Genomic DNA was extracted from blood samples. The coding exons and intronic flanking regions of the RECQL4 (21 exons; NM_004260), C16orf57/USB1 (7 exons; NM_024598.3) and TWIST1 genes (1 exon; NM_000474.3), were amplified using a touchdown protocol (PCR primers and conditions available on request). The small introns of RECQL4 (7, 8, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20) were also sequenced. Polymerase chain reaction (PCR) fragments were purified using the multiscreen Vacuum Manifold system (Millipore, Darmstadt, Germany). Sequencing was performed using an ABI BigDye Terminator Cycle Sequencing kit (v3.1; Applied Biosystems, Foster City, CA) in an ABI 3130 sequencer. Sequence data were analysed with SeqScape v2.7 (Applied Biosystems) and Sequencer v4.1. The pathogenicity of mutations was evaluated by phylogenic studies, single-nucleotide polymorphism (SNP) databases studies and in silico analyses using sift and polyphen programs.
Array-comparative genomic hybridization (CGH) analysis
A targeted oligonucleotide array-CGH study was designed with approximately 180,000 oligonucleotides manufactured by Agilent Technologies, Inc (Santa Clara, CA), covering the entire genome with an average resolution of 15 kb and increased probe density within three selected genes implicated in syndromic poikiloderma (RECQL4, C16orf57 and KIND1) and four genes implicated in syndromic craniosynostosis (FGFR1, FGFR2, FGFR3 and TWIST). A graphical overview was obtained using Genomic Workbench software (v5.0) and ADM-2 statistical algorithms according to sensitivity threshold 6.0 and a moving average window of 0.5 Mb. Mapping data were analysed on the human genome sequence using Ensembl (www.ensembl.org).
Clinical analysis
In order to determine the best indicators for RECQL4 screening, a clinical analysis of patients positive for RECQL4 mutations was first performed, followed by the search for differential diagnoses in negative patients through clinical revision of the diagnosis and/or complementary molecular analyses. The remaining patients were compared to patients with RECQL4 mutations.
Results
RECQL4, C16orf57/USB1 and TWIST sequencing analysis
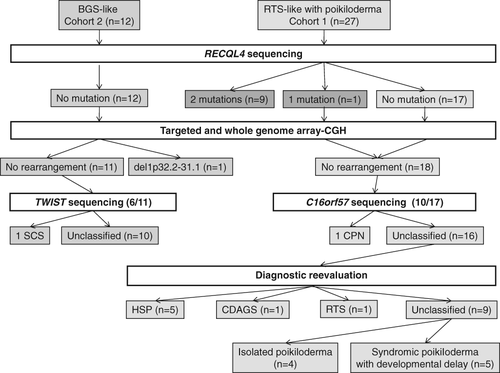
Twelve different RECQL4 mutations were identified in 10/27 patients of cohort 1 (Table 1). Altogether, recurrent p.(Gln757*) and p.(Cys525Alafs*33) mutations were present in 9/19 alleles. Novel mutations were identified (Table 1). No RECQL4 mutations were found in cohort 2. C16orf57/USB1 analysis identified two mutations in one patient [p.(Trp81*) and p.(Tyr89*)] (6; Fig. 2). TWIST1 analysis in six RECQL4-negative BGS patients with craniosynostosis identified one heterozygous causal mutation [p.(Gln3*)] in a 26-year-old female presenting with craniosynostosis and right thumb hypoplasia, associated with a cleft palate, oesophageal atresia, short stature and dysmorphic features.
| RTS patients with two RECQL4 mutations (n = 9) | Patient with one RECQL4 mutation (n = 1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patients | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Sex (male/female) | F | F | M | F | F | F | F | M | F | M |
| Age at referral (years or months) | 38.5y | 6 years | 6 years | 8 years | 16 months | 21 years | 6 years | 12 years | 3 years | 17 years |
| Sibling recurrence | + | −c | − | − | − | − | − | + | + | + |
| Consanguinity | + | − | − | − | − | − | − | + | − | − |
| Facial dysmorphismd | + | + | + | + | + | + | + | + | + | + |
| Skin abnormalities | ||||||||||
| Poïkiloderma | + | + | + | + | + | + | + | + | + | + |
| Facial distribution of poikiloderma | + | + | + | + | + | + | + | + | + | + |
| Scalp alopecia/dysplastic hair | − | + | − | + | − | − | − | − | − | − |
| Eyebrows alopecia | − | NA | + | + | + | + | + | + | − | + |
| Eyelashes alopecia | + | NA | + | + | − | + | − | − | − | + |
| Dental abnormalities | NA | − | − | + | NA | + | − | NA | − | − |
| Growth retardation | + | + | + | + | + | + | + | − | + | − |
| Skeletal abnormalities | ||||||||||
| Thumb agenesis/hypoplasia | − | + | − | + | − | + | − | + | − | − |
| Other radial ray defects | − | + | − | + | − | − | − | + | − | − |
| Patellar hypoplasia | − | NA | NA | + | − | + | NA | NA | NA | − |
| Craniosynostosis | − | − | − | − | − | − | − | − | + | − |
| Other skeletal abnormalities |
Scoliosis Two patellar exostosis |
NA | Short hands and feet | Short hands and feet | − |
Short feet Duplicated left thumb |
Short limbs | Short feet | Short hands | − |
| Digestive disorders | NA | NA | NA | − | Vomiting | Vomiting | Diarrhoea | − | − | − |
| Other visceral malformationsa | − | − |
Inguinal hernia Cryptorchidy |
− | + |
Inguinal hernia Anal anteposition |
− | − | − | − |
| Eyes abnormalities | ||||||||||
| Cataract | − | − | − | − | − | − | − | − | − | − |
| Other eye abnormalities | − | − | − | − | − | − | Right ptosis | − | − | − |
| Cancer | − | − | − | − | − | − | − | − | − | − |
| Intellectual deficiency | − | − | − | − | − | − | − | − | − | − |
| Probable RTS diagnosisb | + | + | + | + | + | + | + | + | + | + |
| Association of at least one of the following features: skeletal abnormalities, growth retardation, digestive abnormalities | + | + | + | + | + | + | + | + | + | − |
| Manifestations limited to skin appendages | − | − | − | − | − | − | − | − | − | + |
| DNA mutations |
c.2269C > T c.2269C > T |
c.1573delT c.1531 T > C |
c.1236G > A c.1573delT |
c.2269C > T c.2545_2546delGT |
c.1913 T > C c.2802G > A |
c.1573delT c.2059-1G > A |
c.2590C > T c.1391-1G > A |
c.2269C > T c.2269C > T |
c.3061C > T c.1573delT |
c.1343_1347delCCACC Unknown |
| Protein changes |
p.(Gln757*) p.(Gln757*) |
p.(Cys525Alafs*33) p. (Cys511Arg) |
p.(Trp412*) p.(Cys525Alafs*33) |
p.(Gln757*) p.(Phe850Profs*33) |
p.(Leu638Pro) p.(Trp934*) |
p.(Cys525Alafs*33) Splice acceptor |
p.(Gln864*) Splice acceptor |
p.(Gln757*) p.(Gln757*) |
p.(Arg1021Trp) p.(Cys525Alafs*33) |
p.(Pro448Argfs*18) Unknown |
- RTS, Rothmund–Thomson syndrome; novel mutations are written in bold characters.
- a Including digestive, genital and cerebral malformations.
- b Probable RTS diagnosis according to Wang and Plon: atypical rash and two of the following clinical signs are present: sparse hair on the scalp, eyebrows and eyelashes, short stature and congenital bone defects (including subtle abnormalities that are visible only on X-rays), dental and nail abnormalities, hyperkeratosis, cataracts, and cancers.
- c Radial malformations in the heterozygous mother.
- d Including brachycephaly (n = 2), round face (n = 1), triangular face (n = 1), high and large forehead (n = 1), large neck (n = 1), ptosis (n = 1), upslanting palpebral fissures (n = 1), enophthalmia (n = 1), thin nose (n = 1), columella extending below alae nasi (n = 2), smooth philtrum (n = 1), large philtrum (n = 1), thin upper lip (n = 2), large ears (n = 1), small ears (n = 1) and low-set ears (n = 1).
Array-CGH analysis
Array-CGH analysis, performed in the patient with only one RECQL4 mutation and in all 29 other patients with negative RECQL4 sequencing analysis only identified a 24.8 Mb deletion in 1p32.2p31.1 (ch1:57817134–82682953;hg19) in a RECQL4-negative patient with intrauterine growth retardation, craniosynostosis leading to turribrachycephaly, facial dysmorphism, right radial and ulna hypoplasia, short first metacarpals and phalanges, bilateral joint luxations with wrist limitations, right lumbar kidney, scoliosis, agenesis of the corpus callosum and intellectual deficiency with no skin lesions.
Clinical analysis
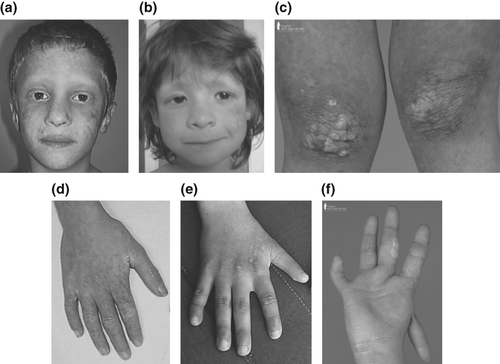
In cohort 1, the patients were analysed depending on whether or not they carried RECQL4 mutations. Detailed clinical data in the 10 patients with RECQL4 mutations are presented in Table 1 (Fig. 1). Besides poikiloderma, all patients had other manifestations of the RTSII spectrum (growth retardation, skeletal manifestations and/or digestive abnormalities), except the patient with only one RECQL4 mutation with a phenotype limited to skin appendages.
Clinical and molecular reevaluation of RECQL4-negative patients led to a diagnosis of HSP in five patients in the presence of tendon contractures and myopathy, CPN in one patient, and CDAGS (craniosynostosis, anal anomalies, and porokeratosis) in one patient in the presence of suspected poikiloderma, alopecia and bone manifestations including craniosynostosis and a sibling presenting with porokeratosis and anal atresia (Fig. 2). Four of the five patients with HSP were molecularly confirmed in the context of the recent identification of the causal gene 7. Among the 10 remaining RECQL4-negative cases, a diagnosis of RTS could not be ruled out despite negative RECQL4 sequencing in one case in the presence of growth retardation and skeletal abnormalities, whereas the others could be classified into two groups: (i) syndromic poikiloderma with developmental delay (n = 5); (ii) isolated poikiloderma when no involvement of organs other than skin appendages (n = 4; Fig. 2). Within the group with syndromic poikiloderma, one patient had juvenile cataract, but it was difficult to raise the diagnosis of RTSI in the presence of developmental delay. Within the group with abnormalities limited to skin appendages, a new entity was probable in a consanguineous family with recurrence in siblings, with total alopecia, normal stature and multiple spinocellular-type skin cancer.
The clinical features of patients of cohort 2 are presented in Table 2.
| BGS patient with one TWIST mutation | BGS patient with abnormal array-CGH | Other BGS patients(n = 10) | |
|---|---|---|---|
| Sex (male/female) | 0/1 | 0/1 | 6/4 |
| Familial history | + | − | 0/10 |
| Consanguinity | − | − | 3/10 |
| Mean age at referral (years) | 26 | 8 | 7 [2.0–21.7] |
| Facial dysmorphism | + | + | 10/10 |
| Craniosynostosis | + | + | 8/10 |
| Type of craniosynostosis | |||
| Coronal | + | NA | 3/7 |
| Other | NA | NA | 3/7 |
| Radial ray malformations | |||
| Thumb agenesis/hypoplasia | + | NA | 8/9 |
| Other radial ray defects | − | + | 7/9 |
| Other skeletal abnormalities | |||
| Patellar hypoplasia | − | NA | 1/3 |
| Joint luxations | − | NA | 2/7 |
| Others | Carpus, metatarsus and phalange abnormalities | Scoliosis | 8/9a |
| Growth retardation | − | NA | 4/8 |
| Skin abnormalities | − | − | 0/10 |
| Intellectual deficiency | − | + | 3/7 |
| Visceral malformations | b | ||
| Digestive malformations | Oesophageal atresia | − | 2/8 |
| Cardiac malformations | − | − | 4/8 |
| Genital malformations | − | − | 1/7 |
| Renal malformations | − | − | 4/8 |
| Cerebral malformations | NA | NA | 2/8 |
| Eye abnormalities | Ptosis | − | 3/8 |
| Others | |||
| Cleft palate | + | − | 1/8 |
- NA, not available.
- a Including five cases with short hands and feet, four cases with joint contractions, four cases with cubital abnormalities, three cases with vertebral abnormalities, and three cases with carpal, tarsal and phalangeal abnormalities.
- b Including anal anteposition (n = 1), oesophageal atresia (n = 1), atrial septal defect (n = 2), ventricular septal defect (n = 1), tetralogy of Fallot (n = 1), complex heart malformation (n = 1), hypospadias (n = 1), renal agenesis (n = 1), hypoplastic kidney (n = 1), kidney ectopia (n = 2), holoprosencephaly (n = 1), thin corpus callosum (n = 1), strabismus (n = 2) and cataract (n = 1).
Discussion
From our series of 39 patients with suspected RTS and BGS referred for RECQL4 screening, we aimed to define the best indicators for RECQL4 sequencing and to search for differential diagnoses in negative patients, in order to help physicians faced with such diagnoses in clinical practice.
We first confirmed that RECQL4 sequencing was not justified in BGS patients without poikiloderma. In order to avoid confusion, the presence of BGS features with poikiloderma would rather be considered a severe presentation of RTS, especially because poikiloderma was not mentioned in the initial definition of BGS 8. We showed that the prescription of array-CGH and TWIST1 sequencing led to higher diagnostic work-up than RECQL4 screening in patients with suspected BGS without poikiloderma 8. Interestingly, another patient with craniosynostosis, abnormal corpus callosum, intellectual deficiency and a 11.2 Mb deletion at the same locus (ch1:58927412–70127412;hg19), has been reported in the Decipher database, suggesting that this locus can be candidate for syndromic craniosynostosis.
Within cohort 1, 25/27 patients met the criteria for possible RTS as defined by Wang and Plon 2 even though only 10 had at least one RECQL4 mutation. These criteria were therefore not appropriate to differentiate positive from negative RECQL4 patients. All of the patients with two RECQL4 mutations had at least one of the three features including growth retardation, skeletal manifestations and digestive abnormalities, and absence of developmental delay.
The search for alternative diagnoses in patients with absence of RECQL4 mutations was a useful strategy. Because CPN could be a differential diagnosis in the presence of poikiloderma, the C16orf57/USB1 gene was screened, leading to CPN diagnosis in one patient with previously undetected neutropenia 6. Because it is now shown that all patients with C16orf57/USB1 mutations have neutropenia, the search for neutropenia should be mandatory in patients with poikiloderma 6. In addition, five patients had features in favour of HSP, in particular tendon contractures and myopathy. HSP has recently been described in a South African family with congenital poikiloderma, hypotrichosis, hypohidrosis, tendon contractures with varus feet, and the causal gene was recently identified as FAM111B 7, 9. In a few cases, the diagnosis of poikiloderma can also be difficult. For example, one patient with suspected poikiloderma was reassigned to CDAGS syndrome after the birth of a sibling, presenting with porokeratosis and anal atresia 10. From this experience, given the high number of genodermatoses presenting with poikiloderma, this pattern of presentation should be considered carefully, with special attention paid to features in favour of CPN or HSP.
In the literature, it is mentioned that RECQL4 mutations could be detected in 60–65% of affected individuals with RTS, suggesting genetic heterogeneity 1. From the retrospective reevaluation, the percentage of convincing negative RTS cases appeared lower in our series (1/10 cases with no RECQL4 mutation, Fig. 2). In the remaining patients, because of partial clinical overlap, ambiguity can be resolved by RECQL4 sequencing until additional progress has been made in the clinical delineation and molecular basis of other syndromes with poikiloderma.
In conclusion, we showed that a significant number of patients reported in the past as RTS with poikiloderma might carry other entities that would benefit from clinical dissection with the help of next generation sequencing tools. This study should lead to a reduction in the prescription of RECQL4 sequencing analysis, but molecular testing remains necessary in cases with overlapping features and undefined diagnosis to ensure appropriate genetic counselling and specific oncosurveillance.
Acknowledgements
The authors thank the association Arc-En-Ciel Wittring and the Regional Council of Burgundy for the financial support of the project, as well as the patients and their families for their participation.




