Founder mutation for Huntington disease in Caucasus Jews
Abstract
Huntington disease (HD), an autosomal dominant disorder involving HTT, is characterized by chorea, psychiatric illness and cognitive decline. Diagnosis and age of onset depend on the degree of expansion of the trinucleotide CAG repeat within the gene. The prevalence of HD is known for Europeans but has not been studied in the Israeli population. Between 2006 and 2011 we diagnosed in our adult genetics clinic ten HD probands, nine of whom were Caucasus Jews (CJ) (Azerbaijani), and one Ashkenazi Jewish. We performed haplotype analysis to look for evidence of a founder mutation, and found that of the nine CJ, eight shared the same haplotype that was compatible with the A1 haplogroup. We calculated the coalescence age of the mutation to be between 80 and 150 years. Ninety percent of our HD patients are CJ, as are 27% of the HD patients in Israel, although the CJ comprise only 1.4% of the Israeli population. Our findings suggest a higher prevalence of HD among CJ compared to the general Israeli population and are consistent with a recent founder mutation. We recommend a higher degree of suspicion for HD in CJ with subtle clinical findings.
Huntington disease (HD [MIM 143100]) is a neurodegenerative autosomal dominant disorder characterized by involuntary movements (chorea), psychiatric illness, memory loss and cognitive decline 1. The protein Huntingtin is encoded by HTT (also called IT15), which is responsible for causing HD. The diagnosis and the age of onset are determined by the degree of expansion of the trinucleotide CAG repeats within the gene 2. The mutated HTT allele in HD patients contains a minimum of 36 CAG repeats. An allele with 27–35 repeats is unstable, and the offspring of such carriers are at risk of expansion to the HD zone, especially when transmitted by the father 3, 4.
The prevalence of HD varies among different populations and is highest in USA and Western Europe; 4–8:100,000 5. The aims of our study were to estimate the prevalence of HD among the various Israeli communities, and to identify the haplotype containing the mutation.
Methods
Clinical data
Following approval from the Rabin Medical Center Institutional Review Board committee, we reviewed the clinical and genetic data of all HD patients referred to the adult genetics clinic of the Rabin Medical Center, Petah Tikva, Israel, which is a large tertiary medical center, between the years 2006 and 2011.
Mutation detection
The mutation was detected by direct amplification of the repeating HTT segment, using the primers: 5′-ATGAAGGCCTTCGAGTCCCTCAAGTCCTTC-3′ and 5′-GGCGGTGGCGGCTGTTGCTGCTGCTGCTGC-3′. The reaction was carried out in a 25 µl reaction volume containing 50 ng of DNA, 13.4 ng of each primer, and 1.5 mM dNTP's, in 1.5 mM MgCl2 PCR buffer, with 1.2 U of Taq polymerase (Bio-Line, London, UK). After an initial denaturation of 5 min at 95°C, 30 cycles were performed (94°C for 30 s, 65°C for 45 s, and 72°C for 30 s), followed by a final extension of 10 min at 72°C. The amplification products were viewed on an ABI 3100 Genetic Analyzer (Applied Biosystems, Grand Island, NY).
Haplotype analysis
Samples were collected from 11 individuals clinically diagnosed with HD and from 31 random control individuals of Caucasus Jewish origin from the general population. DNA was purified from peripheral blood leukocytes using standard protocols.
The haplotype surrounding HTT was defined in the patients and controls by eight polymorphic markers using the ABI 3130 genetic analyzer (Applied Biosystems) with PCR products marked by fluorescent primers. Table 1 shows the primers used.
| Marker | hg19 location (base) | Forward | Reverse |
|---|---|---|---|
| D4S3038 | 10998981100210 | GAAGACCAGCATTCGG | GGTTTAATACACAGTAATTGTTCA |
| 17 × TG | 1881552–1881586 | TCACCCGTGTTCAGTGAGAG | CCACAGAACAGAGCGTCAAA |
| 40 × AC | 1953171–1953251 | CCTGGGATTGAGTTGTCTCC | AAAACAGGCCTAGCACCTCA |
| D4S1614 | 2646545–2646878 | CATCTAGGAGAATCAGTACTTGG | TTACCATGAGCATATTTCCA |
| HTT | 3076408–3245687 | ||
| D4S412 | 3380692–3381012 | ACTACCGCCAGGCACT | CTAAGATATGAAAACCTAAGGGA |
| D4S3023 | 4301335–4301697 | ACCTCACTGGAAACTAAATGG | TGAACAGCAGCGGTCT |
| D4S2925 | 4893183–4893399 | TCAGAAACCCCTACAGGAAA | TTTGATGAGTTATTCGGAGG |
| D4S2285 | 5103989–5104325 | ATGAGCTCCTCTGAGAGG | GGAAAGAGGGCAAGACTC |
To further define the HD haplotype we sequenced eight single nucleotide polymorphisms (SNPs) surrounding HTT in all the HD patients. The SNPs were chosen by their ability to discriminate between the HTT region haplotypes based on Warby et al. 2009 6 (Fig. 1). The SNPs were analyzed by direct sequencing using ABI 3130 genetic analyzer (Applied Biosystems). Phasing was performed manually and therefore was inferred. The common haplotype was phased based on HD102 and HD111, siblings who inherited the CAG expanded haplotype from their HD-affected deceased father and different alleles from their healthy mother, who was not available for genotyping.

Dating the founder mutation and relatedness of ancestors
Dating the founder mutation in Caucasus Jews (CJ) was carried out using the dmle+2.0 software program (www.dmle.org) 7, 8. It was based on the following parameters: (1) genotypes of polymorphic markers in 62 control chromosomes and haplotypes of eight chromosomes carrying the mutation. (2) Map distances between markers and the mutation taken from the human genome working draft GRch37/hg19. (3) The growth rate (r) of the CJ estimated from the equation: T1 = T0e(gr), where T1 is the estimated size of the CJ population in the world at the present time (120,000–140,000) 9-11, T0 is the estimated size of the ancestral population, i.e. ˜10,000–15,000 in the 17th century 12, and g is the number of generations between T1 and T0 assuming 20 years per generation. The calculation yielded a growth rate of 0.118. (4) The estimated allele frequency of the founder HTT mutation in the general CJ population based on the estimated number of CJ HD patients in Israel and a total of 105,000–120,000 CJ in Israel (C. Bram and M. Sicron: personal communication 17 January 2013) 13-15.
Given the strong founder effect previously suggested for CJ 16, 17, we tested for cryptic relatedness among our patients at the whole genome level using the Illumina HumanOmniExpress BeadChip array composed of ˜730 K SNPs (Illumina, San Diego, CA). Genotyping was performed following the manufacturer's directions. Data were evaluated and analyzed using Illumina's genomestudio v2011.1. The recent ancestry was estimated by measuring the total size of identical by descent (IBD) shared segments between all pairs of HD patients when a segment is defined by a minimum of 500 consecutive SNPs in a raw on the OMNI BeadChip providing it is no less than 1 cM as taken from HapMap.
Results
Patients
Between the years 2006 and 2011, 1,400 patients were referred to the adult genetics clinic at our center. A total of 11 patients from 10 different families were diagnosed with HD. Of ten probands, nine were of CJ origin and one was of Ashkenazi Jewish ancestry. Table 2 gives the detailed clinical and genetic data of the patients. The average age of onset of symptoms was 45 years (range: 29–61 years). There were no statistically significant differences between the genders. The average age of genetic diagnosis was 55 years (range: 36–68 years). The number of CAG repeats expanded to between 39 and 50. There was a mean gap of 6.9 years between onset of symptoms and confirmation of the diagnosis (HD).
| Patient ID | M/F | Ethnicity | Age of onset | Age of genetic diagnosis | Delay in diagnosis (in years) | Involuntary movements | Cognitive and memory decline | Psychiatric complaints | Brain MRI | No. of CAG repeats | Family Hx | Parent of HD origin |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HD101 | M | CJ | 49 | 55 | 6 | + | + | Depression | n/a | 42, 16 | + | Mother |
| HD102 | M | CJ | 35 | 44 | 9 | + | + | Aggressive responses | Global cerebral atrophy, symmetric putamen nuclei atrophy, low signal of Globus Pallidus | 45, 17 | + | Father |
| HD103 | F | CJ | 29 | 36 | 7 | + | + | Depression | Cerebral atrophy | 50, 17 | + | Father |
| HD104 | F | CJ | 56–60 | 64 | 4–8 | + | − | Depression and upset mood | Diffuse hyperintense T2 changes in centrum semiovale and hemispheres, compatible with ischemic changes | 40, 18 | + | ? |
| HD105 | M | CJ | 63 | 66 | 3 | + | + | Depression | Moderate dilation of ventricles, mild periventricular | 40, 17 | + | Mother |
| Hyperintensity in flair and T2 | ||||||||||||
| HD106 | M | CJ | 50 | 55 | 5 | + | + | Mood changes, anger | n/a | 42, 24 | + | Mother |
| HD107 | M | CJ | 60 | 65 | 5 | + | + | Mood changes | MRI n/a, brain CT – mild cerebral atrophy | 40, 16 | ? | ? |
| HD108 | F | AJ | 64 | 68 | 4 | + | − | Depression | MRI n/a, brain CT – cerebral atrophy | 39, 15 | + | Father |
| HD109 | F | CJ | 46 | 52 | 6 | + | + | Depression | n/a | 41, 15 | Neg | / |
| HD110 | F | CJ | 34 (Dep – 29, schiz – 34) | 49 | 15 | + | + | Depression, schizophrenia | normal | 43, 18 | Neg | / |
| HD111 (sister of HD102) | F | CJ | 40 | 50 | 10 | + | + | Aggressive responses | n/a | 43, 18 | + | Father |
- AJ, Ashkenazi Jews; CJ, Caucasus Jews; Dep, depression; F, female; HD, Huntington; Hx, history; ID, identification; M, male; MRI, magnetic resonance imaging; schiz, schizophrenia.
Haplotype analysis
Analysis of the patients' haplotypes around HTT revealed that eight of the nine CJ HD patients shared an extended haplotype based on the eight markers; this haplotype was found in 1 of 62 chromosomes of CJ in the general population. The 3.74 Mb shared haplotype is located 1.71 Mb upstream and 1.86 Mb downstream from HTT (Fig. 1). Individual HD108, who is of Ashkenazi Jewish origin, had a different haplotype, as did individual HD107, who is CJ.
The specific HD haplogroups were assessed using eight SNP's and revealed that the shared haplotype is compatible with the HD susceptible A1 haplogroup, although phasing was performed manually and therefore was inferred (Fig. 1). We could not exclude the A4 haplogroup, because all eight patients with the shared haplotype were heterozygous for rs362307 (C/T), meaning that their haplotypes could be compatible with either the A1 or the A4 alleles. However, the A4 allele seems very unlikely, because it would force the ‘T’ allele at rs362307 onto the non-expanded chromosome, making it an unknown haplotype, rather than the common ‘C’ haplotype on non-expanded chromosomes. The CAG-expansion in HD107 and HD108 was assumed to be on the HD A2 haplogroup.
Dating the founder mutation and estimation of relatedness
To date the coalescence age of the founder HTT mutation in the CJ population we used the dmle + 2.0 program based on the parameters described in the methods. On the basis of an estimate of 66 CJ HD patients out of a total of 245 HD patients diagnosed in the two HD diagnostic laboratories in Israel and ˜112,500 CJ in Israel, we calculated the allele frequency of the founder mutation to be 0.000293. However, we know that HD is frequently misdiagnosed 18, 19, and therefore for our calculation we also doubled the actual number of patients to compensate for the under-diagnosis. We estimated that the mutation occurred 4.3–7.6 generations ago, corresponding to 80–150 years ago (Fig. 2).
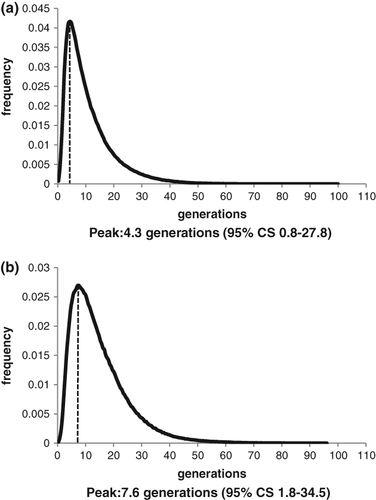
For estimation of relatedness at the whole genome level we studied the total and largest IBD haplotype segments shared by our patients and as inferred from ˜730 K SNPs genotypes available from the BeadChip platform. HD108 showed no evidence of recent ancestry within the last six generations and was actually predicted to be of Ashkenazi ancestry. HD102 and HD111 showed a relatedness pattern compatible with a parent/child or full sibling pattern. All other comparisons suggested the individuals to be first to fifth degree relatives.
Discussion
HD is one of the most devastating neurodegenerative diseases. Its prevalence is highest in the USA and Western Europe (4–8 cases per 100,000 individuals) 5. Jewish immigrants constitute the majority of Israeli society and we expected the prevalence of HD to be equal among the various ethnicities. However, we noted that the CJ HD patients comprise 90% of HD patients in our genetics clinic and 27% of the HD patients in Israel, even though the CJ account for only 1.4% of the Israeli population (the total Israeli population is based on the 2011 Israeli Central Bureau of Statistics). In Israel the CJ are a small and closely related population of 105,000–120,000 who are scattered around all parts of the country, with the majority concentrated mostly in the periphery. The CJ who live in the center of Israel are served by our genetics clinic. In the literature there are reports of a high prevalence of HD in Azerbaijan, which was one of the countries where the CJ's resided during the Diaspora 20; however, we were unable to find a comprehensive molecular study in this population. We conducted this clinical and genetic study in order to expand our knowledge of HD in the CJ population.
Statistics based on CJ population size should be regarded with caution because tracing the demographic characteristics of CJ is a difficult task 21 and there is no official census count. One estimate by Nikolas Witsen put the size of the original CJ population at the end of the 17th century at about 15,000 12. There is evidence that some Jews converted to Islam, and it is probable that many also left the region altogether 21. Taking all these statistics together, we suggest that an approximate figure of 10,000–15,000 was the probable size of the original CJ population at the end of the 17th century, because it is probable that only a part of the population survived or kept their Jewish identity 21.
The clinical characteristics of our HD patients are shown in Table 2. There is a mean of 6.9 years' delay in the diagnosis of HD in our patients; this is probably a result of the frequent misdiagnosis of these patients, which is known to be a problem 18, 19. In one extreme case, patient HD110, there was a 15 year delay between a schizophrenic episode and the diagnosis of HD. This case shows the need for a high level of awareness of the higher prevalence of HD among CJ.
The molecular analysis of the HTT locus in our cohort of HD patients showed a common founder haplotype in 8 of the 10 probands (Fig. 1), a second haplotype in the AJ patient (HD108), and, surprisingly, a third haplotype in another CJ HD patient (HD107). The shared CJ HD haplotype is 3.74 Mb long and HTT is located approximately in the middle. By dating this founder mutation we found that it is of recent origin occurring 80–150 years ago. In a further study for ancestry relatedness among our CJ patients we found them to be first to fifth degree relatives. This is compatible with the suggested demographics of the population and molecularly attests to a strong founder effect, isolation, and the probable high level of inbreeding of the CJ. It is also concordant with the recent origin of the mutation.
It has been suggested that many factors contribute to CAG instability 6. Warby et al. proposed the predisposing haplogroup model, where certain haplotypes surrounding HTT are predisposed to CAG expansion based on genetic cis-elements and trans-factors, whereas other haplogroups are more stable 6, 22. It was shown that the A1 and A2 haplogroups are more prevalent in HD patients, suggesting that chromosomes harboring these haplogroups are more susceptible to CAG-expansion 6. To study this further in our CJ HD patients, we analyzed all the HD patients using SNPs known to have discriminatory power between the various HD haplogroups. In the common CJ haplotype we could not differentiate between the A1 and A4 haplogroups because all the patients were heterozygous for rs362307 (C/T); however, the A1 allele is more probable as it allows the non-expanded chromosome to be a ‘C’ haplotype rather than an unknown allele (Fig. 2). This is also supported by Warby et al. 6, who showed that a cluster of similar haplotypes (haplotype A) is present on 95% of HD chromosomes, where the majority of these (55%) could be classified as the A1 allele and the A4 allele was almost absent from the expanded CAG chromosomes. Both patients HD107 and HD108, who did not harbor the common CJ haplotype, have haplogroup A2, which is the second most common risk allele.
Conclusion
In view of our finding of a founder mutation that causes a higher prevalence of HD among CJ compared to the general population in Israel and the documented average delay of around 7 years to diagnosis, we want to raise awareness among physicians of HD in this unique CJ population. This will enable earlier diagnosis allowing for prenatal evaluation and possible preimplantation genetic diagnosis when desired, and when specific and effective treatment becomes available, early initiation of such treatment.
Acknowledgments
The authors thank Prof. Moshe Frydman and Dr Israela Lerer for their help with the manuscript, Prof. Moshe Sicron for his assistance in calculating the CJ population size and Dr Gabrielle J. Halpern for her editorial assistance.




