Design, Synthesis and Evaluation of Benzimidazole Derivatives as IL-6 Inhibitors and Their Role in Rheumatoid Arthritis
Funding: The authors received no specific funding for this work.
ABSTRACT
Interleukin-6 (IL-6) is a pleiotropic cytokine that plays a major role in the development of Rheumatoid Arthritis (RA). In the present study, benzimidazole and benzene sulfonyl scaffold were identified as pharmacophore by analysis of literature reports and novel small molecule IL-6 inhibitors were designed. These were screened via docking with IL-6 (PDB: 1ALU), then and through Lipinski's rule of 5. Based on docking score, 10 best compounds (4a–4e and 7a–7e) were selected for synthesis and evaluated for IL-6 inhibitory activity in vitro. Compounds 4b and 7b showed the maximum inhibition of IL-6 (87.55% and 82.75%, respectively). These compounds were further explored for anti-arthritic activity in vivo using the Incomplete Freund's Adjuvant Model and by morphological and histopathological studies of the inflamed paw. Compound 4b was significantly more active than compound 7b and both were found to be slightly less active than methotrexate. These findings indicate that a benzimidazole nucleus linked to a benzene sulphonyl moiety may prove to be a useful template for the development of new chemical moieties against RA.
1 Introduction
Rheumatoid Arthritis (RA) is largely controlled by pro-inflammatory cytokines such as Tumor Necrosis Factor Alpha (TNF-α), Interleukin-1β (IL-1β) and Interleukin-6 (IL-6) Brennan, Maini, and Feldmann (1998). These play a crucial role in the pathology of RA by causing the induction of differentiation of helper T-cells Narazaki, Tanaka, and Kishimoto (2017). An increased level of IL-6 accelerates joint destruction by inducing or suppressing the differentiation of osteoclast or osteoblast, respectively. Osteoclast genesis occurs due to the induction of Receptor Activator of Nuclear Factor Kappa-Β Ligand (RANKL) in osteoblasts and fibroblasts by IL-6 or its receptor (IL-R) signals (Pandolfi et al. 2020; Nishimoto 2006). Tocilizumab, an anti-IL-6R antibody, inhibits bone destruction and enhances the repair of bone erosion in RA patients. Even though biologics are more effective than synthetic drugs, their usage has been constrained due to cost, restricted intravenous administration and other limitations (Takeuchi, Yoshida, and Tanaka 2021). Hence, targeting IL-6 with small molecules is a promising strategy in the treatment of RA.
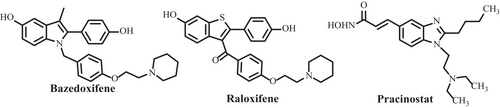
Various compounds targeting IL-6 known in literature can be broadly classified as IL-6 production inhibitors, IL-6 expression inhibitors, IL-6R antagonists and IL-6 signaling inhibitors (Kaur et al. 2020). Lately, a new mode of IL-6 inhibition has been reported by (Fiest and Nasonov 2023) wherein olokizumab is found to inhibit IL-6 by directly binding with it. Bazedoxifene, raloxifene and pracinostat are USFDA-approved clinically significant molecules that suppress IL-6-induced STAT3 phosphorylation in GP130/JAK/STAT3 signaling pathways (Figure 1) (Tian et al. 2019; Luo et al. 2020; Chen et al. 2020). (+)−Madindoline A and B (Figure 2) have been reported to bind directly to IL-6 to block receptor interaction as well as inhibit IL-6 production pathways (Saleh et al. 2005; Yamamoto et al. 2006). Lately, Yoo et al. (2022) have reported benzoxazole derivatives for the treatment of RA due to their ability to inhibit IL-6/STAT3 signaling pathway.
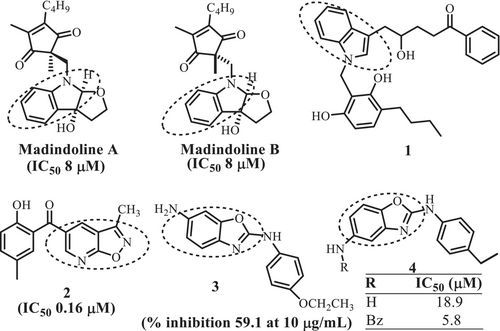
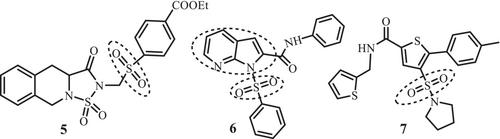
Several other compounds containing indole (1), benzisoxazole (2) and benzoxazole (3–4) rings (Figure 2) have also been reported to target IL-6, which indicates that benzo-fused five-membered heterocyclic-based compounds can afford important pharmacophore for IL-6 inhibition (Kaur et al. 2020). Sulfonamide unit is identified as an important descriptor in anti-inflammatory (celecoxib), antibacterial (sulfasalazine) and cardiovascular (Doburil) drugs (Zhao et al. 2019). Sulfonyl group is also present in compounds 5 and 6 inhibit IL-6 production and signaling mechanisms Keche et al. (2012). Derivatives of 3-(pyrrolidine sulfonyl)-thiophene (7) (Figure 3) have been reported to inhibit the transcriptional activity of STAT3 (EC50 15 μM) in HeLa cells to activate STAT3 signaling via the IL-6/gp130 receptor (Zinzalla et al. 2010).
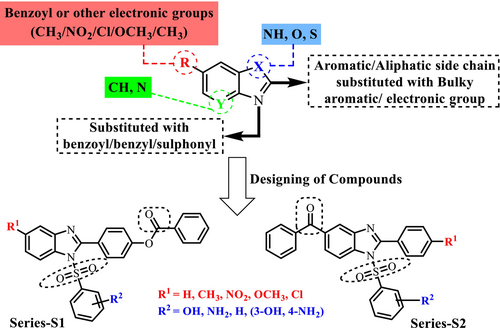
These literal findings envisioned our research to design benzimidazole derivatives as potential IL-6 inhibitors for the treatment of RA. An insightful look into the structures of these known IL-6 inhibitors has revealed that a carbonyl group and a sulfonyl group are responsible for binding to the active site of human IL-6. In the present study, these two pharmacophores were coupled with a benzimidazole nucleus to design two series of compounds S1 and S2 (Figure 4). These were screened via docking into the active site of IL-6 (PDB 1ALU) and via Lipinski's rule of 5. Compounds with the best docking score were synthesized and evaluated for IL-6 inhibitory activity. The compounds that showed > 70% inhibition of IL-6 were further evaluated in vivo using the incomplete Freund's adjuvant model of RA.
2 Results and Discussion
2.1 Docking-Based Screening of Compounds
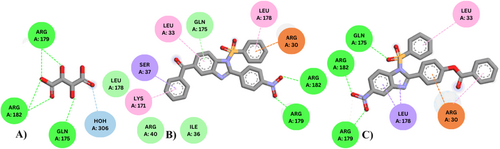
Human IL-6 is a polypeptide containing 185 amino acids that interact with IL-6R (α-chain) and gp130 (β-chain) in the process of molecular recognition and signaling. Tartaric acid (TLA) is associated with human IL-6 on a crystallographic 2-fold axis, representing the stoichiometry of one tartrate bound to two IL-6 molecules. Docking of TLA (the co-crystal ligand) with IL-6 showed good binding interactions through Arg182 and Arg179 with a docking score of −4.7 kcal/mol. Tartrate's carboxyl groups facilitate the binding via direct H-bonding with Arg182 and Arg179. Additionally, the α-hydroxy group of tartrate accepts an H-bond from Arg179 whereas the β-hydroxy group donates an H-bond to the oxygen of Gln175 (Figure 5) (Somers, Stahl, and Seehra 1997). Similar H-bonding and pi-pi interactions were observed between the designed compounds and human IL-6 through the same amino acid residues but with binding affinity better than TLA. Compounds 4a, 4c–4e, 7a, 7c and 7e bind through H-bonds between their carbonyl moiety and Arg179 and Arg182, whereas compounds 7b and 4b bind by H-bonding through nitro (−NO2) group with Arg179 and Arg182 (Figure 5). The sulfonyl (SO2) group of compounds 7c and 7a shows additional H-bond interaction with Gln175, whereas compound 4a shows interaction with Arg30. Molecular Docking data and results of Lipinski Rule of 5 for varied 5-substituted benzimidazole and 5-benzoyl benzimidazole derivatives are given in (Tables S1 and S2), respectively. The detailed interactions visualized after docking are shown in (Table S3).
2.2 ADME Properties
Predicting absorption, distribution, metabolism and excretion (ADME) is an important tool in the early stages of the drug development process. It helps to identify the compounds that are more likely to exhibit satisfactory ADME performance during clinical trials. ADME properties of the designed compounds were predicted by using the SwissADME (2023) online webserver (Keche et al. 2012). It is a quick, accurate and easy-to-use method to estimate properties such as molecular weight (MW), H-bond donor (HBD), H-bond acceptor (HBA), molecular refractivity (MR), topological polar surface area (TPSA), partition coefficient (cLog P) and number of rotatable bonds (NRB). All these parameters for the target compounds selected for synthesis on the basis of docking were found within acceptable limits (Table 1).
| Compound | R | MW (g/mol) | HBD | HBA | MR | TPSA (Å2) | cLogP (o/w) | NRB | |
|---|---|---|---|---|---|---|---|---|---|
| 4a | H | 427.19 | 0 | 5 | 125.64 | 86.64 | 4.92 | 6 | |
| 4b | –NO2 | 472.49 | 0 | 7 | 134.46 | 132.46 | 4.18 | 7 | |
| 4c | –CH3 | 441.21 | 0 | 5 | 133.55 | 86.64 | 5.26 | 6 | |
| 4d | –Cl | 461.15 | 0 | 5 | 130.65 | 86.64 | 5.44 | 6 | |
| 4e | –OCH3 | 457.20 | 0 | 6 | 132.13 | 95.87 | 4.96 | 7 | |
| 7a | H | 456.22 | 0 | 5 | 124.11 | 70.83 | 4.83 | 7 | |
| 7b | –NO2 | 483.50 | 0 | 6 | 132.94 | 123.23 | 4.27 | 6 | |
| 7c | –CH3 | 470.23 | 0 | 4 | 129.08 | 77.41 | 5.18 | 5 | |
| 7d | –Cl | 490.18 | 0 | 4 | 145.21 | 77.41 | 5.35 | 5 | |
| 7e | –OCH3 | 468.52 | 0 | 5 | 130.61 | 86.64 | 4.80 | 6 |
- Note: Acceptable limits: MW < 500 g/mol; HBD < 5; HBA < 10; MR40-130; TPSA20-130; cLogP < 5; NRB < 9.
2.3 Chemistry
The selected target compounds were synthesized using the methods outlined in Schemes 1 and 2. In Scheme 1, 5-substituted o-phenylenediamine (1) was treated with 4-hydroxybenzoic acid in the presence of orthophosphoric acid (OPA) to give various 5-substituted benzimidazoles (2a–2e) (Kaur et al. 2008). Compounds were characterized through IR, 1H-NMR, 13C-NMR and HRMS techniques. In IR spectra of compounds 2a–2e, the CO stretching band (due to -COOH in 4-hydroxybenzoic acid) was absent, which suggests that the coupling of –COOH group with o-phenylenediamine has occurred to form benzimidazole nucleus. In 1H-NMR spectra, a broad singlet due to OH was observed at δ 10.04–8.42 whereas the aromatic protons were detected at δ 8.30–5.59. Compounds 2a–2e were further treated with benzoyl chloride in the presence of NaOH to obtain intermediates 3a–3e. The disappearance of the band at 3581–3610 cm−1 (due to –OH in 2) and the appearance of a CO stretch at 1737–1744 cm−1 in IR spectra of compounds 3a–3e ascertained the formation of an ester linkage. Lastly, intermediates 3a–3e were stirred with benzene sulfonyl chloride in the presence of triethylamine (TEA) and dichloromethane (DCM) to obtain the target compounds 4a–4e (Holiyachi et al. 2016). The disappearance of the NH band in IR spectra marked the formation of these compounds. In Scheme 2, variedly substituted benzoic acids (5a–5e) were refluxed with 3, 4-diaminobenzophenone in OPA to obtain respective substituted 2-phenyl-5-benzoyl benzimidazole derivatives (6a–6e), which were then sulfonylated with benzene sulfonyl chloride using the reaction conditions as in Scheme 1 to get the target compounds 7a–7e. All target compounds (4a–4e and 7a–7e) showed asymmetric and symmetric SO2 bands at about 1326–1330 and 1139–1140 cm−1 in IR spectra indicating the presence of sulfonyl linkage. The physicochemical and spectral data of all compounds is given in Supporting Information.
2.4 Biological Evaluation
2.4.1 In Vitro IL-6 Inhibitory Activity
The target compounds (4a–4e and 7a–7e) were evaluated for in vitro IL-6 inhibitory activity using a Mouse IL-6 GenLisa ELISA kit and ELISA Reader at 450 nm. All compounds exhibited anti-IL-6 activity > 60% (Table 2). The compounds bearing electron-withdrawing groups (NO2 and Cl) were more active than those bearing electron-donating groups (OCH3 and CH3). Among these, the nitro-substituted compounds (4b and 7b) showed the maximum inhibitory activity (87.55% and 82.75%, respectively). The methyl-substituted compounds (4c and 7c) were even less active than the corresponding unsubstituted compounds (4a and 7a). The activity of all compounds was lower than that of dexamethasone (taken as a standard drug).
| Compound | % Inhibition of IL-6 at 10 μM |
|---|---|
| 4a | 63.55 ± 0.84 |
| 4b | 87.55 ± 0.92 |
| 4c | 62.11 ± 0.89 |
| 4d | 74.33 ± 1.09 |
| 4e | 70.56 ± 1.14 |
| 7a | 63.06 ± 1.74 |
| 7b | 82.06 ± 1.10 |
| 7c | 60.89 ± 1.11 |
| 7d | 73.75 ± 1.14 |
| 7e | 68.63 ± 0.96 |
| Dexamethasone | 99.04 ± 0.66 |
2.4.2 Safety Study
Determining the dosage of a medication that does not have adverse effects at the highest acceptable level has proven beneficial in the early stages of the drug development process. According to Borgert, Fuentes, and Burgoon (2021) tests at doses 10–100 times higher than the expected dose can be used to determine this level in terms of NAOEL (No Observed Adverse Effect Level). Compounds 4b and 7b were found to have significant therapeutic efficacy at 1 mg/kg. Consequently, a first non-clinical risk evaluation was conducted with a dosage of 10 mg/kg. There were no negative effects seen in animals administered compounds 4b and 7b at this dosage. The behavior of the treated animals remained normal and no death was noted throughout 48 h.
2.4.3 In Vivo Antiarthritic Activity
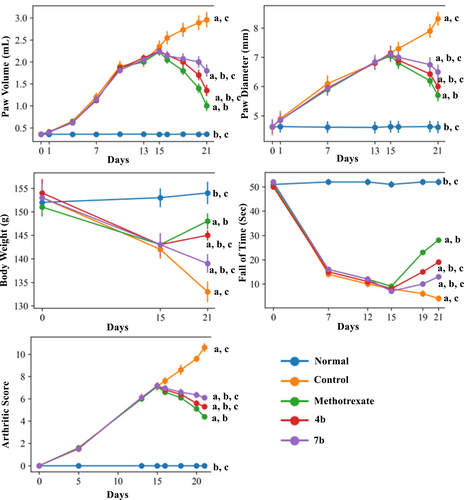
Among all target compounds, 4b and 7b showed the maximum in vitro IL-6 inhibitory activity. Therefore, these were further evaluated at the dose of 1 mg/kg for antiarthritic activity in vivo taking methotrexate as the standard drug (1 mg/kg) in Incomplete Freund's Adjuvant ICFA-induced arthritis model. The activity was evaluated in terms of paw thickness, paw volume, body weight, fall time as a measure of locomotor activity and arthritic score. Subplantar administration of adjuvant in animals caused a significant increase in paw thickness and volume (expressed in terms of volume and diameter), loss of body weight and difficulty in locomotor activity with respect to normal animals on day 15. Treatment of disease control animals with 4b and 7b (once daily) during days 16–21 resulted in significant gain in body weight of the animals as well as a significant reduction in paw volume and diameter in comparison to the disease control (Figure 6). The locomotor activity was evaluated using a Rota-Rod device wherein rats were placed for 1 min on the rotating rod of the Rota-Rod apparatus. The time taken for the rat to fall from the roller during the 1 min period was recorded. Both test compounds (4a and 7b) showed significant and gradual improvement in fall time as compared to the disease control (Figure 6). Arthritic score was assigned on the basis of morphological features of arthritis, such as redness, swelling and erythema through visual criteria as reported in the literature Kaur, Singh, and Silakari (2017). Both the target compounds showed significant improvement in arthritic score as compared to the disease control (Figure 6). However, improvement in all these pathological indications by compounds 4b and 7b was less than that brought about by methotrexate. Nevertheless, these findings revealed that these compounds have the potential to treat rheumatoid arthritis.
2.4.4 Morphological and Histopathological Studies
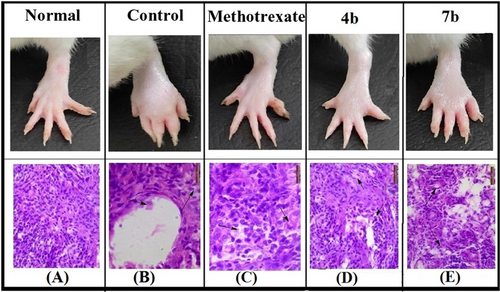
Synovium was observed to be normal in size, the cartilage was entire and smooth and the specimens lacked any signs of inflammation in the paw of normal animals (Figure 7A). Treatment of animals with IFA (disease control animals) severely inflamed the paw and showed multiple pathological features (Figure 7B). Dermis and subcutaneous tissue were severely inflamed in disease-control animals. Chronic inflammatory cells like lymphocytes and macrophages were found present in the inflammatory exudate. Additional multi-nucleated giant cells were also observed. With the invasion of mononuclear cells, there was synovial fibroblast hyperplasia or proliferation (pannus formation). Variable loss of cartilage was noticed in the joint surfaces. Additionally, the periosteum showed an increase in fibrous tissue. One sample also revealed osteoclastic activity, bone resorption or lysis. Treatment of disease-control animals with methotrexate, significantly reduced the inflammation and improved the histopathological features of the dermis and hypodermis, though synovium showed some mononuclear cell infiltration (Figure 7C). Treatment of disease control animals with 4b or 7b significantly decreased the inflammatory response induced by IFA (Figure 7D,E). Synovium, skin and cartilage surfaces of these treated groups were also found to be significantly improved. A close analysis suggests that compound 4b exhibits anti-arthritic activity better than compound 7b.
2.5 Molecular Dynamics Simulation
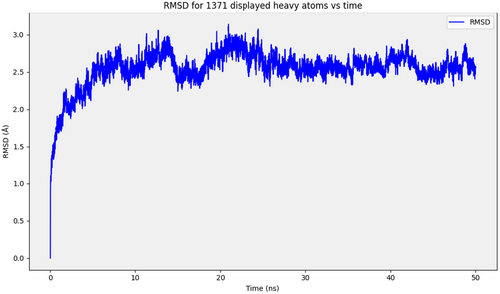
The protein-ligand complex stability, binding mode and potential interactions within the binding site of 1ALU were analyzed for the most potent compound (4b) by molecular dynamic simulation studies for a 50 ns period. Complexes with thermodynamically stable conformations are produced by interactions with respect to time. It was found that cation-pi stacking interactions between Arg179 and the aromatic carbon atoms of 4b remained constant. The docking contacts were maintained during dynamic simulations. Important interactions vanished while some hydrophobic and pi-pi interactions with Gln175, Arg179 and Phe74 with a frame score < 50% are sustained. The percent frame score for 4b which includes the essential amino acids is presented in (Table S4). Overall, the 4b-1ALU complex was stable as indicated by minimal RMSD fluctuations (Figure 8).
2.6 Structure Activity Relationship (SAR)
An analysis of the structure and activity of compounds reveals that a NO₂ substituent exhibits the best binding affinity and the maximum IL-6 inhibition indicating a strong positive effect on both binding affinity and biological activity. A Cl or OCH₃ group also enhances the activity, but is less effective than NO₂. Conversely, a CH₃ substituent shows the lowest IL-6 inhibition and docking scores, even less than a hydrogen substituent, indicating a negative impact on both. These patterns are consistent across both series, highlighting the significant influence of electronic and steric effects of substituents on the effectiveness of compounds.
3 Conclusion
Two series of compounds were designed using the pharmacophores identified for IL-6 inhibition. All designed compounds were docked with IL-6 (PDB: 1ALU). The compounds capable of binding with active sites of the protein were selected for predicting their ADME properties and for synthesis. The compounds showed good ADME properties and followed Lipinski's rule of 5. All synthesized compounds exhibited > 50% IL-6 inhibition at a 10 μM concentration in vitro. Compounds 4b and 7b were found to be the most potent inhibitors of IL-6 (87.54% and 82.55% inhibition, respectively). The in vivo study revealed that compound 4b was more effective in decreasing inflammation compared to compound 7b. The molecular dynamics studies suggested that the complex of compound 4b with the protein target was stable and displayed minimal RMSD fluctuations. The compound was found safe as it shows no observable adverse effects at 10 mg/kg dose level. The compound 4b can be taken as a lead molecule for further optimization of structural descriptors for the development of novel potent anti-RA drugs.
4 Materials and Methods
4.1 Computer and Software
Molecular modeling was carried out on an HP Pavilion G-4 with an AMD processor (4.00 GHz), 8GB RAM, 64-bit and Windows 10 as the operating system. ChemBio Draw Ultra 15.0 was utilized for drawing chemical structures. AutoDock Vina 4.2.6 and Biovia Discovery Studio 2022 were used for molecular docking studies and visualization, respectively. Molecular dynamics studies were performed using Cresset Flare 8.0.
4.2 Molecular Docking and ADME Studies
4.2.1 Preparation of Ligand and Protein
2D structures of designed compounds were drawn in ChemBioDraw Ultra 15.0, converted to 3D structures and MMF94 force field was applied to minimize their energy. Subsequently, these were converted into. pdbqt file format for molecular docking with IL-6 (PDB: 1ALU). The binding site was defined according to the reported co-crystallized ligand. The grid box of size was X = 30, Y = 38 and Z = 30 with the coordinates −7.5087, −12.8277 and 0.0565, respectively to perform docking. The PDB structure (1ALU) was downloaded from the Protein Data Bank. It was refined and prepared using UCSF Chimera and Biovia Discovery Studio. After the preparation of the protein, the designed compounds were docked into its hot spot site.
4.2.2 ADME Properties
ADME properties were predicted using the SwissADME (2023) online web server that helped to estimate the absorption, distribution, metabolism and excretion (ADME) properties of the compounds. These properties include Partition Coefficient (log P), Molecular Weight (MW), H-Bond Donor, H-Bond Acceptor (HBA), Topological Polar Surface Area (TPSA), Molecular Refractivity (MR) and Number of Rotatable Bonds (NRB).
4.3 Chemistry
4.3.1 General
Each reaction was monitored with thin-layer chromatography (TLC) using pre-coated aluminum plates. Completion of reactions and purity of the compounds was ascertained by observing the developed TLC plate in a UV chamber at both short and long wavelengths. All intermediates and compounds were characterized using IR, NMR and HRMS spectral analysis. IR spectra were recorded as KBr pellets and vibrational frequencies were presented as wavenumbers in cm−1. For NMR spectra were recorded in DMSO-d6 with tetramethyl silane as an internal standard and chemical shifts were reported as δ values. High-resolution mass spectral (HRMS) data were generated in positive electrospray ionization mode (+ESI) wherein each compound was identified as [M+H]+ ions at m/z values corresponding to its precise mass.
4.3.2 Synthesis of Compounds
The selected compounds were synthesized using Schemes 1 and 2. For the synthesis of compound 2, a mixture of o-phenylenediamine (2.5 mmol) and 4-hydroxybenzoic acid (5 mmol, 0.69 g) was refluxed with orthophosphoric acid (OPA) in a 50 mL round bottom flask. After the completion of the reaction (4–6 h), the reaction mixture was poured into an excess of cold water and made alkaline with dilute ammonia solution. The resultant precipitated product was filtered, washed with cold water and recrystallized from hot methanol to obtain the pure product [20]. Subsequently, a solution of compound 2 (1 mmol) in 10% NaOH (10 mL) was stirred in a round-bottom flask on a magnetic stirrer for about 30 min. Benzoyl chloride (3 mmol, 0.5 mL) was added dropwise to the stirred solution. Stirring was continued until the product precipitated out. The resulting product was filtered, washed thoroughly with water to remove any impurities and dried to obtain the corresponding product (Mann and Saunders 1975). Finally, for the synthesis of target compound 4, triethylamine (1.2 mmol) was added dropwise to a cooled (0°C) solution of 3 (0.36 mmol) in dichloromethane (10 mL). After stirring for 10–15 min, benzene sulfonyl chloride (0.36 mmol) was added in portions over 30 min. The mixture was stirred at room temperature for 8–10 h. Upon completion, the reaction mixture was washed thrice with 2 M HCl, followed by water and small portions of saturated NaHCO₃ solution. The organic layer was dried over anhydrous sodium sulfate and the solvent was recovered under vacuum to obtain the corresponding final compound 4. For synthesis of target compound 7, compound 6 was prepared from benzoic acid (5a) or p-substituted benzoic acid (5b–5e) using the method as that used for 2, which was finally sulfonated using the method as that used for 4.
4.4 In Vitro IL-6 Inhibition Activity
Spleen from untreated mice that were used as control groups for other approved investigations at the institution was removed. These were perfused with 3 mL of RPMI-1640 medium supplemented with 5% fetal bovine serum (FBS) and then centrifuged at 1250 rpm for 10 min at 4°C. The supernatant was discarded, the splenocyte pellet was mixed with lysis buffer (150 mL, pH 7.4, 0.15 M ammonium chloride, 1 μM potassium bicarbonate and 0.1 μM EDTA) and incubated at room temperature for 5 min with occasional gentle shaking. The mixture was centrifuged again for 5 min at 4°C, and the supernatant was discarded. The splenocytes were resuspended in fresh RPMI-1640 medium and centrifuged at 1250 rpm for 10 min. The resulting splenocyte pellet was isolated and re-suspended in new RPMI-1640 medium to form a splenocyte suspension with a cell density of about 2 × 108 cells/mL. To each well of a 96-well microtiter plate, 100 μL each of the splenocyte suspension, lipopolysaccharide (LPS, 2.5 μg/mL) and the target compound/dexamethasone solution were added. The plate was incubated at 37°C for 48 h. IL-6 levels were quantified using an ELISA reader using an IL-6 ELISA kit by following the manufacturer's instructions. Both dexamethasone and the target compounds were tested at concentrations of 10 μM to determine their percent inhibition (Kaur and Bansal 2022; Rott et al. 2003; Dorato and Engelhardt 2005; Hock et al. 2020).
4.5 Safety Study
As suggested by (Dorato and Engelhardt 2005) and (Borgert, Fuentes, and Burgoon 2021), a non-clinical risk assessment of target compounds 4b and 7b was conducted in terms of No Observed Adverse Effect Level (NOAEL). For this assessment, a dose of 10.0 mg/kg body weight was administered orally to Wistar rats (n = 5). The control group received regular saline. The risk evaluation focused on mortality, behavioral abnormalities and adverse or toxic effects observed within 48 h post-injection.
4.6 In Vivo Anti-Arthritic Activity Using IFA-Induced Arthritis Model
The anti-arthritic activity of compounds 4b and 7b was evaluated using the Incomplete Freund's Adjuvant (IFA)-induced arthritis model (Bendele 2001). Wistar rats of either sex, weighing more than 155 g, were procured from the animal house of Punjabi University, Patiala. The experimental protocol (107/GO/ReBi/S/99/CPCSEA/2021-9) was approved by the Institutional Animal Ethics Committee. The animals were divided into 5 groups (I-V) with five animals in each group. The group I was taken as a normal control, received the normal diet and remains untreated. Arthritis was induced in groups II–V. To induce arthritis, each rat was administered an intradermal injection of 0.1 mL of IFA into the plantar surface of the left hind paw. Arthritis became clinically apparent on day 15. The group II (vehicle-treated) was taken as disease control that received normal saline post-day 15. The groups III–V (drug-treated) were treated with methotrexate (standard drug), 4b and 7b, respectively from day 16 to day 21 with a dose of 1 mg/kg (p.o.). Methotrexate is a widely used disease-modifying antirheumatic drug that is clinically used in the treatment of RA. Therefore, it was selected as a standard drug in the current study for comparisons. The severity of arthritis was assessed by measuring paw edema and the arthritic score. Paw volume was measured using a mercury plethysmometer up to the ankle joint on days 0, 1, 3, 5, 8, 13, 15, 16, 18 and 21 in both the vehicle and drug-treated groups. The arthritic score was determined using a visual scoring system, with each paw graded from 0 to 4 based on the degree of erythema, edema and joint deformity. A score of 0 indicated no swelling or erythema; 1 indicated slight erythema or swelling of one toe or finger; 2 indicated slight erythema and swelling of two toes or fingers; 3 indicated slight erythema and swelling of the ankle or wrist; and 4 indicated complete erythema and swelling of the toes or fingers, with the ankle or wrist unable to bend. The maximum possible arthritic index after the sum scores of all four legs, was 16. Arthritic scores were recorded on days 13, 15, 16, 18, 20 and 21 in both the vehicle and drug-treated groups (Kaur and Bansal 2022).
4.7 Statistical Analysis
All the results were expressed as mean ± SD and analysed using two-way ANOVA followed by the Bonferroni post hoc test. Statistical analyses were performed with GraphPad Prism software and a p-value of < 0.05 was considered to indicate statistical significance.
4.8 Molecular Dynamics Simulation
The protein-ligand complex stability, binding mode and potential interactions within the binding site of 1ALU were analyzed for the most potent compound 4b by molecular dynamic simulation studies using AMBER 18 (Flare 8.0, Cresset package) Salomon-Ferrer, Case, and Walker (2013); Wang et al. (2004); Jakalian, Jack, and Bayly (2002). Using the explicit solvent water model, a solvent system was constructed around the protein-ligand complex and the box's dimensions (10 × 10 × 10 Å) were maintained in an orthorhombic form. Afterwards, the energy of the complex was minimized by setting the minimum energy tolerance as 0.25 kcal/mol and equilibrating each complex for 200 ps. The simulation was performed at a time step of 1 fs and 50 ns as recording time. The stability of the complex was analyzed by RMSD and compared before and after simulation. Frames were captured every 4 ps and saved a trajectory. A total of 5000 frames were generated during the simulation of each complex. The default parameters for MD simulations were used. The charges of ligands binding to the catalytic site were determined using AMI-BCC and GAFF techniques.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that supports the findings of this study are available in the Suporting Information of this article.




