ELF1 suppresses autophagy to reduce cisplatin resistance via the miR-152-3p/NCAM1/ERK axis in lung cancer cells
Lifeng Zhao, Xiangsheng Wu, and Zhiwen Zhang are co–first authors.
Abstract
Resistance to chemotherapeutic drugs limits the efficacy of chemotherapy in non–small cell lung cancer (NSCLC). Autophagy is an essential mechanism which involves in drug resistance. Our previous research has revealed that miR-152-3p represses NSCLC progression. However, the mechanism of miR-152-3p in autophagy-mediated chemoresistance in NSCLC remains unclear. Cisplatin-resistant cell lines (A549/DDP and H446/DDP) were transfected with related vectors and subjected to cisplatin, autophagy inhibitor, activator, or extracellular signal–regulated kinase (ERK) activator. Flow cytometry, CCK8 and colony formation assays were performed for testing apoptosis and cell viability. The related RNAs or proteins were detected by qRT-PCR or Western blot. Chromatin immunoprecipitation, luciferase reporter assay or RNA immunoprecipitation were used for validating the interaction between miR-152-3p and ELF1 or NCAM1. Co-IP verified the binding between NCAM1 and ERK. The role of miR-152-3p in cisplatin resistance of NSCLC was also validated in vivo. The results showed that miR-152-3p and ELF1 were decreased in NSCLC tissues. miR-152-3p reversed cisplatin resistance by inhibiting autophagy through NCAM1. NCAM1 promoted autophagy through the ERK pathway and facilitated cisplatin resistance. ELF1 positively regulated miR-152-3p level by directly interacting with miR-152-3p promoter. miR-152-3p targeted NCAM1 to regulate NCAM1 level and then affected the binding of NCAM1 to ERK1/2. ELF1 inhibited autophagy and reversed cisplatin resistance through miR-152-3p/NCAM1. miR-152-3p inhibited autophagy and cisplatin resistance of xenograft tumor in mice. In conclusion, our study revealed that ELF1 inhibited autophagy to attenuate cisplatin resistance through the miR-152-3p/NCAM1/ERK pathway in H446/DDP and A549/DDP cells, suggesting a potential novel treatment strategy for NSCLC.
Abbreviations
-
- CCL2
-
- C-C motif chemokine ligand 2
-
- DNMT1
-
- DNA methyltransferase 1
-
- ERK
-
- extracellular signal–regulated kinase
-
- NCAM1
-
- neural cell adhesion molecule 1
-
- NSCLC
-
- non–small cell lung cancer
-
- OAS1
-
- oligoadenylate synthetase 1
-
- TNFAIP8
-
- tumor necrosis factor ɑ–induced protein 8
1 INTRODUCTION
Lung cancer is the most common cause of cancer-related deaths worldwide.1 Currently, cisplatin has become one of the “first-choice” drugs used to treat various solid tumors, including lung cancer.2 However, chemoresistance results in treatment failure, which limits the clinical utility of cisplatin for non–small cell lung cancer (NSCLC) patients.3 Hence, deeper understanding of the drug resistance mechanisms can contribute to developing strategies for overcoming chemoresistance.
Autophagy is a catabolic process that captures and degrades cellular waste in lysosome,4 which is a physiological mechanism for cell survival effectively applied by cancer cells to cause drug resistance.5 Our previous study has demonstrated that DNMT3B regulates the proliferation of A549 cells through the miR-152-3p/NCAM1 pathway.6 Importantly, miR-152-3p is implicated in modulating autophagy in acute myeloid leukemia.7 DNA methyltransferase 1 (DNMT1) represses mitophagy and exacerbates heart failure through miR-152-3p.8 Besides, circRNA1615 modulates autophagy by miR-152-3p in cardiomyocytes of myocardial infarction.9 However, whether miR-152-3p could affect cisplatin resistance by regulating autophagy needs to be studied in NSCLC.
Neural cell adhesion molecule 1 (NCAM1) is considered a signaling receptor that impacts cellular migration, proliferation, apoptosis, and survival.10 In lung cancer, knockdown of NCAM1 inhibited cell migration, invasion, and autophagy. At the same time, NCAM1 promoted the development of leukemia and drug resistance. Literature has reported that extracellular signal–regulated kinase (ERK) signaling is closely related to autophagy in cancer.11, 12 Therefore, we hypothesized that NCAM1 could regulate autophagy via ERK signaling, thus affecting cisplatin resistance in NSCLC.
E74, like ETS transcription factor 1 (ELF1), has been reported as a transcription factor that is involved in the progression of diverse cancers.13, 14 ELF1 transcriptionally modulates downstream molecular through binding to their promoters to facilitate their transcriptions, participating in cancer development. For example, ELF1 enhances the expression of C-C motif chemokine ligand 2 (CCL2) through binding to its promoter in nasopharyngeal carcinoma cells.15 ELF1 binds to the promoter of 2′-5′-oligoadenylate synthetase 1 (OAS1) and promotes the transcriptional response to interferon-β.16 Furthermore, ELF1-activated long noncoding RNA (lncRNA) CASC2 inhibits cisplatin resistance via miR-18a in NSCLC.17 TCGA online prediction shows that ELF1 expression is low in lung cancer, which is associated with poor prognosis. Moreover, bioinformatics analysis in TransmiR predicts that ELF1 can regulate miR-152-3p. Hence, we wondered whether ELF1 could interact with miR-152-3p to modulate cisplatin resistance in NSCLC. In our research, cisplatin-resistant A549/DDP and H446/DDP cells were employed to explore whether ELF1 modulates autophagy to affect cisplatin resistance via the miR-152-3p/NCAM1/ERK pathway in NSCLC.
2 MATERIALS AND METHODS
2.1 Cell culture
Human NSCLC cells (A549 and H446) and the cisplatin-resistant cells (A549/DDP and H446/DDP), obtained from FengHui Biotechnology were cultured in RPMI 1640 medium (Thermo Fisher) supplemented with 100 U/mL streptomycin/penicillin and 10% fetal bovine serum (Thermo Fisher). Cells were incubated at 37°C in a humidified environment with 5% CO2 in a humidified incubator. For the generation of cisplatin-resistant cells, in brief, A549 and H446 cells were cultured with 5 μM of cisplatin (PHR1624, Sigma) for 3 days, followed by 3 days without cisplatin (3 days on/off cisplatin treatment) for a total of 3 months. Afterward, 20 μM of cisplatin was used to treat the cells for the next 3 months with the same procedures (3 days on/off cisplatin treatment).
2.2 Cell transfection
miRNA negative control (mimics NC, 5′–UAGCCACGGUUGUGUAAAGUCUG–3′), miR-152-3p mimics (5′–UCAGUGCAACUGACAGAACUUGG–3′), inhibitor negative control (inhibitor NC, 5′–UCGCUUGGUGCAGGUCGGGAA–3′), miR-152-3p inhibitor (5′–CCAAGUUCUGUCAUGCACUGA–3′), overexpression negative control (oe-NC), pcDNA3.1-NCAM1, and pcDNA3.1-ELF1 were purchased from RiboBio. A549/DDP and H446/DDP cells (3 × 103 cells/well) were planted in 96-well plates. When grown to 70% cell confluence, cells were transfected with plasmids using Lipofectamine 2000 reagent (Invitrogen) in accordance with the manufacturer's guidelines for 48 hours.
2.3 Drug treatments
Cells were then treated with cisplatin (20 μg/mL, Sigma) for 24 hours following transfection. Autophagy activator rapamycin (10 μM, Sigma), autophagy inhibitor 3-methyladenine (10 mM, Sigma), or ERK1/2 activator (5 μM, Calbiochem) was added into cells with cisplatin treatment. Afterward, cells were collected for further analysis.
2.4 Immunofluorescence
After fixing by paraformaldehyde (4%), cells were permeabilized by Triton-X (0.5%), followed by blocking in bovine serum albumin (2%). They were incubated in primary LC3 antibody (ab128025, Abcam) at 4°C overnight before incubation with secondary antibody (ab150077) for 1 hour. The nucleus was stained by DAPI. A confocal microscope (Olympus) was applied for fluorescence detection.
2.5 Western blot
RIPA lysis buffer (Beyotime) was employed for cell lysis. Then, protein concentrations were tested by a protein assay kit (Beyotime). Following separation on sodium dodecyl sulphate-polyacrylamide gel (SDS-PAGE), protein specimens were removed onto nitrocellulose membranes (Sigma). Membranes were incubated in primary antibodies at 4°C overnight prior to incubation with secondary antibodies. ECL reagent (Thermo) was used to detect protein bands. The primary antibodies included anti-Beclin1 (ab207612, Abcam), anti-p62 (ab109012, Abcam), anti-LC3 (ab128025, Abcam), anti-ERK1/2 (#9102, Cell Signaling), anti-p-ERK (#9101, Cell Signaling), and anti-NCAM1 (ab237708, Abcam). The protein levels were normalized to GAPDH (ab9485, Abcam).
2.6 CCK8 assay
After transfection, cells were exposed to cisplatin (0, 0.25, 0.5, 1, 2, 4, 8, 16, and 32 μg/mL) for 24 hours with or without other drugs. Then, cell viability was detected by adding 10 μL of CCK8 reagent with CCK8 assay. Following incubation for 1.5 hours at 37°C, absorbance was recorded on a microplate reader (Bio-Rad) at 450 nm. Dose-response curves were established for determining IC50 value.
2.7 Flow cytometry
Annexin V-PE/7-AAD apoptosis kit (Sigma) was used to evaluate cell apoptosis. Flow cytometry (Beckman Coulter) was applied for quantifying apoptosis, after cells had been stained by Annexin V-PE and 7AAD. Cells were categorized into late-apoptotic cells (Q4), viable cells (Q3), early-apoptotic cells (Q2), and dead cells (Q1).
2.8 Colony formation assay
Cells (1 × 103 cells/well) were seeded in a six-well plate after being subjected to the indicated treatments. The clones were rinsed by PBS after incubation at 37°C for 12 days. Next, they were incubated in 1.0% crystal violet overnight after fixation in 4% formaldehyde. The formed clones containing more than 50 cells were manually counted.
2.9 Co-immunoprecipitation
Cells were lysed using RIPA buffer before incubation with anti-IgG (ab172730, Abcam), anti-NCAM1 (ab237708, Abcam), or anti-ERK1/2 (ab184699, Abcam) for 2 hours. Afterward, the samples were incubated for 1 hour in protein A/G Plus agarose beads. After rinsing with RIPA buffer, immunoprecipitates were determined using Western blot.
2.10 Tissue microarray (TMA)-based immunohistochemistry (IHC) and chromogenic in situ hybridization (CISH)
Tissue microarray containing 150 dotted adjacent and tumor tissues from 75 NSCLC patients was acquired from Biochip. miR-152-3p and ELF1 were measured using IHC, with specific RNA-CISH Probe (Ribobio) and rabbit anti-ELF1 polyclonal antibody (ABSIN, abs148134), respectively. All experiments were approved by the Ethics Committee of The Affiliated Hospital of Youjiang Medical University for Nationalities.
2.11 qRT-PCR analysis
Total RNA was obtained by TRIZOL reagent (Invitrogen). Then, a reverse-transcription kit (Takara) was applied for synthesizing cDNA in accordance with the manufacturer's instruction. RNA expressions were detected using qRT-PCR with SYBR Premix Ex Taq II (Takara) and gene-specific primers. ABI 7500 real-time PCR system (Life Technologies) was used to conduct the PCR reactions. RNAs expressions were determined with 2−ΔΔCT method relative to U6 or GAPDH.
2.12 Luciferase reporter assay
To generate pGL3-ELF1 mutant (MUT) and pGL3-ELF1 wild type (WT), or pGL3-NCAM1-MUT and pGL3-NCAM1-WT, the fragments of ELF1 or NCAM1 containing the forecast mutant or wild-type binding sites of miR-152-3p were cloned into pGL3 vector (Promega). Cells (2 × 105 cells/well) were planted and cultured in a six-well plate overnight. Next, they were transfected with miR-152-3p inhibitor, inhibitor NC, miR-152-3p mimics, or mimics NC, along with pGL3 constructs by Lipofectamine 2000 (Invitrogen) for 48 hours. After transfection, a dual-luciferase reporter assay system kit (Promega) was employed to measure relative luciferase activity.
2.13 RNA immunoprecipitation (RIP) assay
EZMagna RIP kit (Millipore) was employed to perform RIP in accordance with the manufacturer's directions. Briefly, cells were lysed in lysis buffer before centrifugation. The supernatant was incubated with control IgG (ab18443, Abcam) or Ago2 antibody (67934-1-Ig, Proteintech) conjugated with protein A/G beads overnight. The isolated immunoprecipitated RNAs were purified before measuring by qRT-PCR. The supernatant was used as input positive control.
2.14 Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation assay kit (Beyotime) was applied for performing ChIP assay in H446/DDP and A549/DDP cells. First, for the cross-linking of protein-DNA complexes, cells (1 × 107) were incubated for 15 minutes in 1% formaldehyde. Next, they were sheared on ice by ultrasound pulses. Afterward, anti-ELF1 (sc-133,096, Santa Cruz) or control IgG (ab18443, Abcam) was used for immunoprecipitation in chromatin specimens. Protein A-sepharose was supplemented and incubated at room temperature for 1 hour. Following de-crosslinking, DNA was purified and detected by qRT-PCR.
2.15 Xenograft tumor model
Male nude mice (BALB/c, 6 weeks old) were obtained from Hunan SJA Laboratory Animal Co., Ltd. They were maintained in sterile conditions, with a relative humidity of 40%-60%, a temperature of 26°C-28°C and a 12 hours light/12 hours dark cycle. Mice were housed with no limitation to food and water. The animal experiment was authorized by the Animal Ethical Committee of the Affiliated Hospital of Youjiang Medical University for Nationalities (No. YYFY-LL-2022-77). Approximately 5 × 106 H446/DDP or A549/DDP cells were resuspended with PBS (200 μL) prior to subcutaneously injection into mice. Then, mice were intraperitoneally administered with 5 mg/kg of cisplatin every week for 3 weeks. The mice were injected with 50 nM miR-152-3p agomir every 5 days. The tumors were measured every 4 days to assess tumor volume according to the formula: volume = length × width2/2. At 25 days, the nude mice were sacrificed by CO2 asphyxia, and tumor tissues were collected.
2.16 Immunohistochemistry
Tumor tissues were fixed in formalin, followed by paraffin embedding, and sectioned at 4 μm thickness. After antigen removal, sections were incubated in H2O2 (3%) to eliminate endogenous peroxidase activity. Sections were incubated in 5% bovine serum albumin (Thermo Fisher), followed by incubation with primary antibodies, including anti-NCAM1 (ab237708, Abcam) and anti-Ki67 (ab16667, Abcam), overnight at 4°C. Afterward, secondary antibodies (ab6721, Abcam) were added and incubated at 37°C for 1 hour, followed by staining with diaminobenzidine and counterstaining with hematoxylin. The slices were observed under a microscope (Olympus).
2.17 TUNEL staining
The apoptosis detection in tumor tissues was carried out by a TUNEL apoptosis kit (Beyotime). In brief, after fixation in 4% paraformaldehyde, tissue sections were permeabilized by 0.3% Triton X-100 in PBS. Next, they were blocked in peroxidase blocking buffer. Slices were incubated for 1 hour with TUNEL reaction mixture, followed by coloring with diaminobenzidine and counterstaining with hematoxylin. TUNEL-positive cells were observed under a microscope (Olympus).
2.18 Statistical analysis
Statistical analysis was carried out in Prism (GraphPad). The data were exhibited as mean ± standard deviation (SD). One-way ANOVA or Student's t test were employed to analyze significant differences among multiple groups or in two groups, respectively. p < 0.05 was considered statistically significant.
3 RESULTS
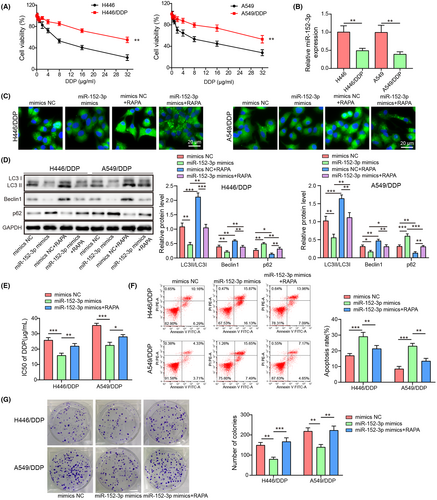
3.1 miR-152-3p inhibited autophagy to reduce cisplatin resistance in A549/DDP and H446/DDP cells
To explore whether miR-152-3p was implicated in autophagy and cisplatin resistance, the drug-resistant cell lines were first screened out by CCK8 assay. Cell viability in H446 or A549 cells decreased more significantly than in A549/DDP or H446/DDP cells with the increased concentrations of cisplatin (Figure 1A). The optimal concentration of cisplatin was 20 μg/mL, which was selected for the subsequent experiments. Compared with those in H446 or A549 cells, miR-152-3p levels in A549/DDP or H446/DDP cells were reduced after cisplatin treatment (Figure 1B). In cisplatin-resistant cells without cisplatin treatment, miR-152-3p levels were decreased, compared with the parental cells (Figure S1A). H446/DDP and A549/DDP cells were transfected with miR-152-3p mimics or mimics NC, respectively, with or without treatment with RAPA. Compared with the mimics NC group, RAPA promoted LC3 expression and miR-152-3p mimics inhibited LC3 expression in DDP-resistant cells (p < 0.05) (Figure 1C). RAPA elevated LC3II/LC3I and Beclin1 levels and decreased p62 level, whereas upregulation of miR-152-3p induced a decrease in LC3II/LC3I and Beclin1 and an increase in p62 (P < 0.05) (Figure 1D). However, RAPA reversed the effects induced by miR-152-3p mimics (P < 0.05) (Figure 1C,D). The autophagy activity of cisplatin-resistant cells without cisplatin treatment was higher than that of parental cells (Figure S1B). With the treatment of cisplatin, miR-152-3p overexpression decreased the IC50 of cells, while the addition of rapamycin promoted cell IC50, illustrating that miR-152-3p reduced cisplatin resistance; however, this impact was dismissed by RAPA (p < 0.05) (Figure 1E). Similarly, miR-152-3p overexpression cells exhibited increased apoptosis and decreased colony number, whereas these effects were reversed by RAPA (p < 0.05) (Figure 1F,G). Thus, we concluded that miR-152-3p repressed cisplatin resistance by inhibiting autophagy in NSCLC.
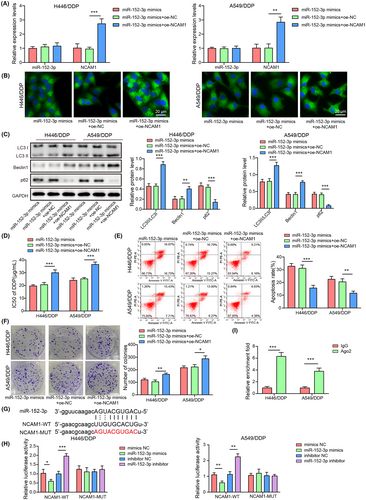
3.2 miR-152-3p suppressed autophagy via NCAM1 to reduce cisplatin resistance
For exploring the potential mechanism of miR-152-3p involved in autophagy-mediated cisplatin resistance, NCAM1 overexpression vector (oe-NCAM), overexpression negetive control (oe-NC), or miR-152-3p mimics were transfected into cells. NCAM1 overexpression did not affect miR-152-3p expression, but increased NCAM1 mRNA level (Figure 2A). Upregulation of miR-152-3p inhibited autophagy (Figure 1C,D), whereas NCAM1 overexpression promoted autophagy and reversed miR-150-3p mimics regulatory effects (p < 0.05) (Figure 2B,C). Besides, miR-152-3p overexpression reduced cisplatin resistance, whereas the effect was overturned by overexpression of NCAM1 (p < 0.05) (Figure 2D). miR-152-3p overexpression exhibited elevated apoptosis and decreased colony number, which were reversed after NCAM1 overexpression (p < 0.05) (Figure 2E,F). Compared with the parental cells, NCAM1 expression was increased (Figure S1C) in cisplatin-resistant cells. Bioinformatics predicted the potential binding sites between miR-152-3p and NCAM1 (Figure 2G). miR-152-3p mimics decreased and miR-152-3p inhibitor increased the luciferase activity in cells cotransfection with NCAM1-WT (p < 0.05) instead of NCAM1-MUT (p > 0.05) (Figure 2H). Anti-Ago2 RIP assay to identify the interaction of miR-152-3p with NCAM1 showed that NCAM1 is a target of miR-152-3p (p < 0.05) (Figure 2I). These outcomes illustrated that miR-152-3p inhibited autophagy to reduce cisplatin resistance of NSCLC cells via targeting NCAM1.
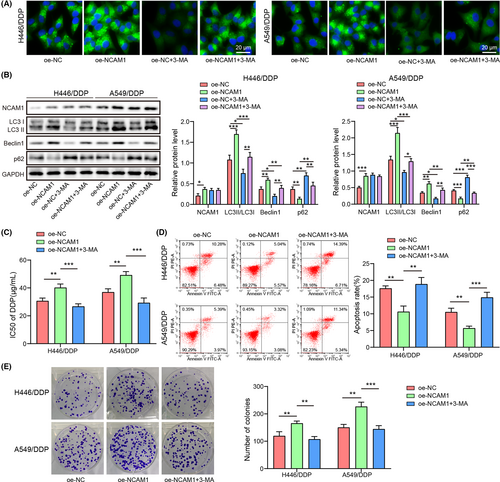
3.3 NCAM1 promotes autophagy to elevate cisplatin resistance
To validate whether NCAM1 was involved in autophagy mediated-cisplatin resistance, oe-NCAM or oe-NC was transfected into cells with or without autophagy inhibitor treatment. Compared with the oe-NC group, autophagy inhibitor inhibited LC3 expression, while NCAM1 overexpression triggered LC3 increase (p < 0.05) (Figure 3A). Autophagy inhibitor decreased LC3II/LC3I ratio, and Beclin1 and increased p62, whereas NCAM1 overexpression upregulated LC3II/LC3I ratio and Beclin1 and downregulated p62 (p < 0.05) (Figure 3B). Moreover, autophagy inhibitor reversed the effects induced by NCAM1 overexpression (p < 0.05) (Figure 3A,B). Additionally, autophagy inhibition dismissed the elevated cisplatin resistance caused by overexpression of NCAM1 (p < 0.05) (Figure 3C). NCAM1 overexpression inhibited apoptosis and increased colony number, which were reversed after autophagy inhibition (p < 0.05) (Figure 3D,E). Taken together, NCAM1 elevated cisplatin resistance of NSCLC cells by promoting autophagy.
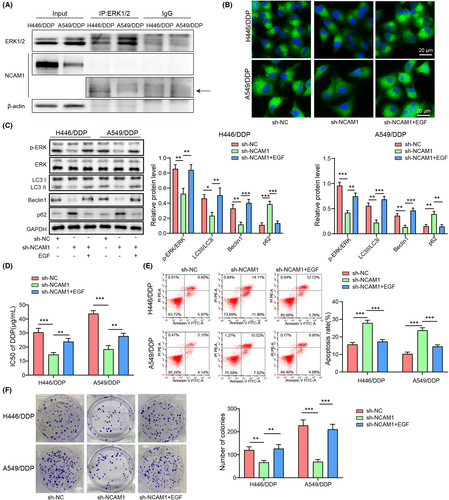
3.4 NCAM1 targeted ERK1/2 to promote autophagy, thereby enhancing cisplatin resistance
For validating the relationship between NCAM1 and ERK1/2, co-immunoprecipitation was performed. The results showed the combination between NCAM1 and ERK1/2 in A549/DDP and H446/DDP cells (Figure 4A). To further verify the involvement of ERK1/2 signaling, sh-NCAM1 or sh-NC was transfected into cells with or without ERK1/2 activator (EGF) treatment. EGF abolished NCAM1 knockdown–induced LC3 decrease (p < 0.05) (Figure 4B). sh-NCAM1 reduced p-ERK/ERK, LC3II/LC3I, and Beclin1 and elevated p62, which were reversed by the administration of EGF (p < 0.05) (Figure 4C). Besides, NCAM1 knockdown reduced cisplatin resistance, while the effect was reversed after EGF treatment (p < 0.05) (Figure 4D). NCAM1-knockdown cells exhibited enhanced apoptosis and reduced colony number, which were dismissed by EGF (p < 0.05) (Figure 4E,F). These data proved that NCAM1 promoted autophagy to elevate cisplatin resistance via interacting with ERK1/2.
3.5 ELF1 positively regulated miR-152-3p expression
Using TMA-based IHC and CISH, we tested ELF1 and miR-152-3p levels in collected paraffin-embedded tissue samples from 75 NSCLC patients. Compared with the matched noncancerous tissues, miR-152-3p and ELF1 were decreased in tissues from NSCLC patients (p < 0.05) (Figure 5A). Compared with H446 and A549 cells, ELF1 expression was decreased in H446/DDP and A549/DDP cells (Figure S1C). The potential binding relationship between miR-152-3p promoter and ELF1 was predicted by bioinformatics analysis (Figure 5B). Overexpression of ELF1 increased luciferase activity in cells cotransfected with miR-152-3p-WT (p < 0.05), whereas the luciferase activity in cells cotransfected with miR-152-3p-MUT had no significant change (p > 0.05) (Figure 5C). Moreover, CHIP analysis found that miR-152-3p promoter closely interacted with ELF1 in NSCLC cells (Figure 5D). ELF1 overexpression triggered an increased miR-152-3p, with respect to the oe-NC group. Conversely, compared with the sh-NC group, miR-152-3p was reduced after inhibition of ELF1 (p < 0.05) (Figure 5E). The results confirmed that ELF1 bound to the promoter region of miR-152-3p to positively modulate the expression of miR-152-3p.
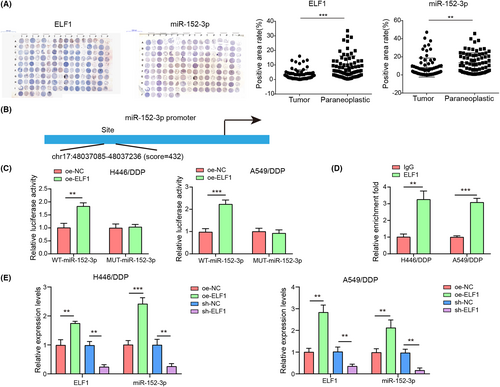
3.6 Overexpression of ELF1 inhibited autophagy and cisplatin resistance, which was abolished by miR-152-3p silencing
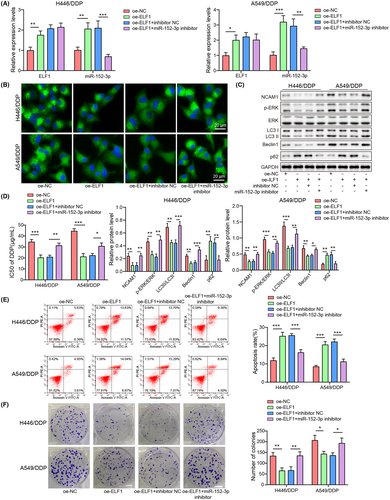
To study whether ELF1 affected autophagy-mediated cisplatin resistance by regulating miR-152-3p, inhibitor NC, miR-152-3p inhibitor, oe-NC, or oe-ELF1 was transfected into cells. miR-152-3p and ELF1 mRNA levels were increased after ELF1 overexpression, while miR-152-3p was decreased, but ELF1 level was not affected by miR-152-3p knockdown (Figure 6A). In comparison with control and oe-NC groups, miR-152-3p was decreased but ELF1 expression was not affected in the oe-NC + miR-152-3p inhibitor group (Figure S1D). Inhibition of miR-152-3p abolished ELF1 overexpression–caused inhibition of LC3 expression (p < 0.05) (Figure 6B). Overexpression of ELF1 downregulated NCAM1, p-ERK/ERK, LC3II/LC3I, and Beclin1, as well as upregulated p62, which were dismissed after miR-152-3p silencing (p < 0.05) (Figure 6C). ELF1 overexpression reduced cisplatin resistance, while inhibition of miR-152-3p could reverse this effect (p < 0.05) (Figure 6D). ELF1-overexpression cells exhibited increased apoptosis and decreased colony number, whereas these effects were overturned following miR-152-3p knockdown (p < 0.05) (Figure 6E,F). Collectively, ELF1 suppressed autophagy-mediated cisplatin resistance through upregulating miR-152-3p in NSCLC.
3.7 miR-152-3p inhibited autophagy and cisplatin resistance of xenograft tumor in vivo
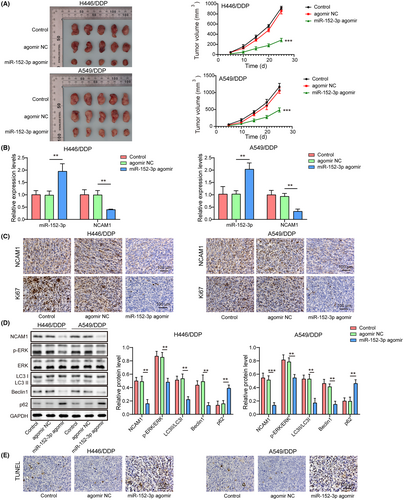
To investigate the role of miR-152-3p in cisplatin resistance in lung cancer in vivo, a xenograft tumor model was established in nude mice. In mice that were injected with miR-152-3p agomir, tumors exhibited less cisplatin resistance than those in the control and agomir NC groups (p < 0.05) (Figure 7A), suggesting that cisplatin resistance was suppressed by miR-152-3p in vivo. miR-152-3p was elevated while NCAM1 mRNA was downregulated in tumor tissues that were injected with miR-152-3p agomir (p < 0.05) (Figure 7B). Immunohistochemistry assay revealed that NCAM1 and Ki67 protein were both downregulated in tumor tissues administrated by miR-152-3p agomir (Figure 7C). In the miR-152-3p agomir group, LC3II/LC3I ratio and Beclin1 were reduced but p62 was elevated; besides, p-ERK/ERK and NCAM1 were decreased (p < 0.05) (Figure 7D). Cell apoptosis was promoted in tumors by miR-152-3p agomir through TUNEL staining (Figure 7E). Collectively, miR-152-3p repressed autophagy and cisplatin resistance of lung cancer in vivo.
4 DISCUSSION
Cisplatin resistance is the leading cause of treatment failure in NSCLC patients.18 We discovered that ELF1 overexpression inhibited autophagy to suppress cisplatin chemoresistance via the miR-152-3p/NCAM1/ERK pathway in NSCLC cells.
Defects in autophagy play an essential role in drug resistance in lung carcinomas following cisplatin treatment.5 In lung cancer, several miRNAs can regulate autophagy activity, thus affecting drug resistance.19 The participation of miR-152-3p in chemosensitivity has been studied in gastric cancer,20, 21 breast cancer,22 and glioblastoma.23 LINC01089 acts as a ceRNA for miR-152-3p to repress NSCLC development.24 Our previous study has demonstrated that miR-152-3p inhibits cell proliferation and invasion of A549 cells via regulation of NCAM1.6 Here, we discovered that miR-152-3p inhibited autophagy and suppressed cisplatin chemoresistance in A549/DDP and H446/DDP cells. Furthermore, autophagy activator rapamycin could overturn the inhibited autophagy and cisplatin chemoresistance induced by miR-152-3p in vitro, indicating that miR-152-3p suppressed drug chemoresistance through inhibition of autophagy. Importantly, we also observed that miR-152-3p inhibited autophagy and cisplatin resistance of xenograft tumors in vivo. It has been reported that miR-152 inhibits doxorubicin resistance in xenografts of breast cancer cells.25 Our study was the first to confirm that miR-152-3p was functionally responsible for autophagy-mediated cisplatin chemoresistance.
As previously shown, elevation of NCAM1 by miR-141-3p represses cell migration in ameloblastoma.26 Perturbation of NCAM1 causes cell differentiation or death, as well as sensitizes leukemic blasts to genotoxic agents.27 Our study revealed that upregulation of NCAM1 reversed the effects induced by miR-152-3p; moreover, autophagy inhibitor overturned the promoted autophagy and cisplatin chemoresistance caused by NCAM1. Our data indicated that miR-152-3p inhibited autophagy to suppress cisplatin chemoresistance via NCAM1 in NSCLC cells with cisplatin resistance. miR-152-3p reduced the NCAM1 level of xenograft tumors in nude mice. Thus, our study uncovered a new function of the miR-152-3p/NCAM1 axis on chemoresistance regulation in NSCLC.
The ERK signaling pathway plays an essential role in the progression of cancer.28 Activated autophagy is associated with the increased ERK signaling in lung cancer.29 The induction of autophagy mediated by ERK activation can resist anticancer activities in lung cancer cells.30 Through our research, we found the binding of NCAM1 with ERK. NCAM1 could activate the ERK signaling pathway, thus promoting autophagy and elevating cisplatin resistance of lung cancer cells with cisplatin-resistance. Furthermore, miR-152-3p reduced p-ERK of xenograft tumors in vivo.
A previous study has revealed that ELF1 enhances transcription of lncRNA LUCAT1 to elevate its expression, which facilitates the progression of choroidal melanoma.31 ELF1 is discovered to be a promoter of lncRNA CASC2 and increase its level in cisplatin-resistant NSCLC.17 ELF1 is recruited to tumor necrosis factor ɑ–induced protein 8 (TNFAIP8) promoter, thereby facilitating TNFAIP8 transcription, which promotes the chemoresistance of acute myeloid leukemia.32 Interestingly, we confirmed a previously undescribed function for ELF1 in transcriptional modulation of miR-152-3p, which positively modulated its expression. Importantly, our data represented that ELF1 activated miR-152-3p expression through targeting its promoter sites.
In summary, the novel role of ELF1 was identified in autophagy-mediated chemoresistance via the miR-152-3p/NCAM1/ERK axis in NSCLC, holding potential to be a therapeutic strategy for cisplatin-resistant NSCLC.
ACKNOWLEDGMENTS
We would like to give our sincere gratitude to the reviewers for their constructive comments.
FUNDING INFORMATION
This work was supported by the Science and Technology Department of Guangxi Zhuang Autonomous Region (grant nos. 2019JJA140044 and 2019JJA140084).
CONFLICT OF INTEREST STATEMENT
All authors have agreed with the presented findings, contributed to the work, and declared no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by an Institutional Reviewer Board. All experiments were approved by the Ethics Committee of The Affiliated Hospital of Youjiang Medical University for Nationalities.
INFORMED CONSENT
Informed consent was obtained from study participants.
REGISTRY AND THE REGISTRATION NO. OF THE STUDY/TRIAL
N/A.
ANIMAL STUDIES
Animal experiments were authorized by the Animal Ethical Committee of The Affiliated Hospital of Youjiang Medical University for Nationalities.
Open Research
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article.




