Predictive value of EGFR mutation in non–small-cell lung cancer patients treated with platinum doublet postoperative chemotherapy
Abstract
The mutation status of tumor tissue DNA (n = 389) of resected stage II-III non-squamous non–small-cell lung cancer (Ns-NSCLC) was analyzed using targeted deep sequencing as an exploratory biomarker study (JIPANG-TR) for the JIPANG study, a randomized phase III study of pemetrexed/cisplatin (Pem/Cis) vs vinorelbine/cisplatin (Vnr/Cis). The TP53 mutation, common EGFR mutations (exon 19 deletion and L858R), and KRAS mutations were frequently detected. The frequency of the EGFR mutation was significant among female patients. Patients with an EGFR mutation-positive status had a significantly shorter recurrence-free survival (RFS) time (24 mo vs not reached) (HR, 1.64; 95% CI, 1.22-2.21; P = .0011 for EGFR mutation status). Multivariable analysis identified both the pathological stage and EGFR mutation status as independent prognostic factors for RFS (HR, 1.78; 95% CI, 1.30-2.44; P = .0003 for disease stage; and HR, 1.57; 95% CI, 1.15-2.16; P = .0050 for EGFR mutation status). This study demonstrated that the EGFR mutation has either a poor prognostic or predictive impact on a poor response to postoperative chemotherapy with platinum doublet chemotherapy for stage II-III Ns-NSCLC patients. This result supports a role for mandatory molecular diagnosis of early-stage Ns-NSCLC for precision oncology and signifies the importance of adjuvant for the 3rd generation tyrosine kinase inhibitor rather than platinum-based chemotherapy. This study is registered with the UMIN Clinical Trial Registry (UMIN 000012237).
Abbreviations
-
- 95% CI
-
- 95% confidence interval
-
- CCP
-
- comprehensive cancer panel
-
- Cis
-
- cisplatin
-
- DFS
-
- disease-free survival
-
- EGFR
-
- epidermal growth factor receptor
-
- FFPE
-
- formalin-fixed paraffin-embedded
-
- HR
-
- hazard ratio
-
- Ns-NSCLC
-
- non-squamous non–small-cell lung cancer
-
- OS
-
- overall survival
-
- Pem
-
- pemetrexed
-
- RFS
-
- recurrence-free survival
-
- TKIs
-
- tyrosine kinase inhibitors
-
- Vnr
-
- vinorelbine
1 INTRODUCTION
Patients with early-stage non–small-cell lung cancer (NSCLC) are operable, but a significant proportion of patients experience recurrence. Adjuvant chemotherapy for early-stage NSCLC patients is the current standard of treatment and is associated with an approximate 5% survival benefit at 5 y.1, 2 The JIPANG study was a randomized phase III study of pemetrexed/cisplatin (Pem/Cis) vs vinorelbine/cisplatin (Vnr/Cis) for completely resected stage II-IIIA non-squamous NSCLC (Ns-NSCLC).3 This phase III study did not meet the primary endpoint (recurrence-free survival, RFS), but Pem/Cis had a similar efficacy to Vnr/Cis, with better tolerability.
Several molecular alterations have been defined as “driver mutations” in NSCLC. These are targets for tyrosine kinase inhibitor. Approximately 30%-40% of NSCLC patients in Asia and 10%-15% of NSCLC patients in the USA and Europe have EGFR mutations.4 Patients with the EGFR mutation are sensitive to EGFR-TKIs, which block cellular growth signaling pathways. EGFR-sensitizing mutations, such as exon 19 deletions (Ex19Del) and the exon 21 point mutation, L858R, are predictive markers for treatment with EGFR-TKIs in patients with advanced NSCLC. EGFR-TKIs are the standard of care for EGFR mutation-positive advanced lung cancer, and a third-generation EGFR-TKI, osimertinib, is now available as a first-line treatment.
Conversely, adjuvant treatment with EGFR-TKIs has not been available for operable NSCLC patients with an EGFR mutation. The ADAURA trial showed that the DFS of patients treated with osimertinib was significantly longer than that of patients treated with a placebo among resected EGFR-mutant NSCLC patients with stage IB to IIIA disease, consistent with the results of the CTONG 1104 and EVAN trials.5-7 Therefore, the prognostic value of EGFR mutation remains unclear for operable NSCLC patients.8-10 Post-trial therapy with EGFR-TKIs is considered to contribute to a better clinical outcome. Therefore, the prognosis and effect of platinum-containing regimens is difficult to analyze in EGFR mutation-positive NSCLC patients.
Biologically, EGFR-sensitizing mutations are strong drivers and may exert a high malignant potential.11 We hypothesized that EGFR-mutant NSCLC patients may have a poor prognosis. We then planned an exploratory biomarker study (JIPANG-TR) to identify predictive and prognostic biomarkers through amplicon deep sequencing-based mutation profiling of tumor tissues. Amplicon deep sequencing is a powerful technology for analyzing somatic mutations in formalin-fixed, paraffin-embedded (FFPE) tumor samples. We previously reported that cisplatin plus pemetrexed is highly effective (long RFS) for patients with high tumor mutation burden levels, which is a predictive biomarker for immune checkpoint inhibitors.12 The JIPANG-TR study, which was open to stage II-III Ns-NSCLC patients with and without EGFR mutations, is an interesting prospective cohort for examining the prognostic or predictive values of platinum doublet chemotherapy for EGFR mutation-positive NSCLC patients. In this study, we analyzed the association between somatic mutations and clinical outcome focusing on single nucleotide variants. We also discuss the prognostic values of mutations, focusing especially on EGFR mutations.
2 MATERIALS AND METHODS
2.1 Clinical specimens and outcome
In total, 389 (48.4%) of the 804 stage II-III Ns-NSCLC patients in the JIPANG study were enrolled in the JIPANG-TR study between March 2012 and August 2016 at each institute (Figure 1). All the patients provided written informed consent to participate in the study (JIPANG-TR), including the collection of tumor tissue for analysis. The clinical outcomes of the JIPANG study (jRCTs041180023) were RFS (primary endpoint) and OS.3 OS was defined as the time from randomization until death from any cause. RFS was defined as the time from randomization until disease recurrence or death, whichever occurred first.
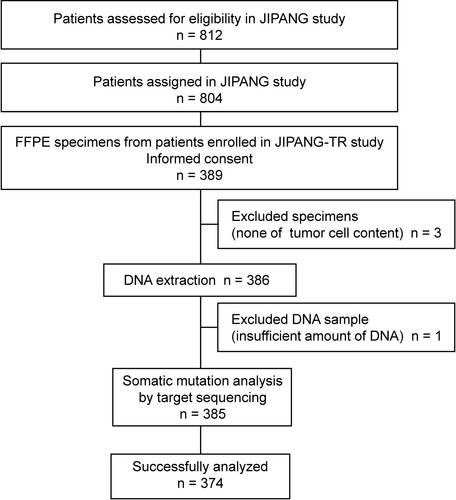
This study was designed as a prospective and exploratory study aimed at characterizing somatic mutations in tumor tissues and comparing the tumor mutation status and RFS with the use of cisplatin-based adjuvant therapy (UMIN000012237). This study was conducted in compliance with the Helsinki Declaration and the Ethical Guidelines for Medical and Health Research Involving Human Subjects by the Japanese government. The study was also approved by the ethics committee of each participating institute.
2.2 Tissue processing
Tumor tissues were obtained during resection and were pathologically confirmed as Ns-NSCLC. The collected FFPE tumor specimens (n = 389) were used for histological review, and only those containing sufficient tumor cells (at least 10%) as revealed by hematoxylin-eosin staining were subjected to nucleic acid extraction (Figure 1). DNA was isolated from these tissues using an AllPrep DNA/RNA FFPE Kit (Qiagen). The quality and quantity of the nucleic acid were verified using a NanoDrop 2000 device and PicoGreen dsDNA Reagent (all from Thermo Scientific).
2.3 Next-generation sequencing
A targeted DNA library comprising approximately 1.2 Mb of the coding regions of 409 genes for panel sequencing was constructed using an Ion AmpliSeq CCP (Thermo Fisher Scientific) in accordance with the manufacturer’s recommended protocol. Briefly, 40 ng of DNA were subjected to multiplex PCR amplification using an Ion AmpliSeq Library Kit 2.0 and an Ion AmpliSeq CCP (Thermo Fisher Scientific), covering all exons in 409 genes. After multiplex PCR, Ion Xpress Barcode Adapters (Thermo Fisher Scientific) were ligated to the PCR products, which were then purified using Agencourt AMPure XP beads (Beckman Coulter). The purified libraries were pooled and then sequenced using an Ion Torrent S5 instrument and an Ion 550 Chip Kit (all from Thermo Fisher Scientific). DNA sequencing data were accessed through the Torrent Suite ver. 5.10 program (Thermo Fisher Scientific). Reads were aligned against the hg19 human reference genome, and variants were called using Variant Caller ver. 5.10. The raw variant calls were filtered with a depth of coverage of <19, quality score of <100, and synonymous variants and were manually checked using the integrative genomics viewer (IGV, Broad Institute). Germline mutations were excluded using the Genome Aggregation Database (gnomAD [>0.1%], ExAC [>0.1%]) and the Human Genetic Variation Database.
2.4 Statistical analysis
Patients were classified based on the EGFR mutation status of the tumor tissues. For biomarker analyses of each somatic mutation, the predictive and prognostic values were assessed by comparing the RFS of each arm (Pem/Cis and Vnr/Cis) in the JIPANG-TR study. JMP (ver. 14.0, SAS Institute) and GraphPad Prism software (ver. 8, GraphPad Software Inc) were used for the statistical analysis. A Cox proportional hazards regression model was applied to perform univariate analyses. The relations between mutation status and patient characteristics were evaluated using the two-sided Fisher exact test. Kaplan-Meier curves were used to estimate survival, and the log-rank test was used to compare times to events between groups. P-values of < .05 were considered statistically significant.
3 RESULTS
3.1 Correlation of somatic mutations with clinical outcomes
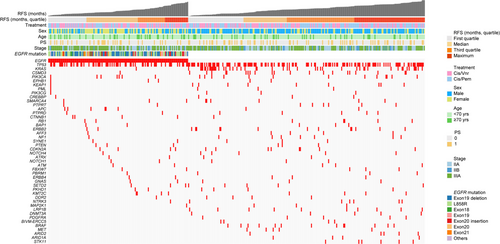
Somatic non-synonymous mutations in FFPE tissue samples (n = 374) were successfully analyzed using targeted deep sequencing (Figure 1). Mutations of TP53 (185/374, 49.5%), EGFR mutations (139/374, 37.2%) and KRAS mutations (51/374, 13.6%) were frequently identified in the 374 samples (Figure 2), as reported previously.13, 14 The exon 19 deletion [Ex19Del] (51/139, 36.7%) and L858R (46/139, 33.1%) mutations were most common EGFR mutations.
RFS and OS were estimated with the Kaplan-Meier method and survival differences were assessed with the log-rank test. The median RFS and OS of this study population (n = 374) were 52.6 mo and not reached, respectively. (Figures 3A and 4A).
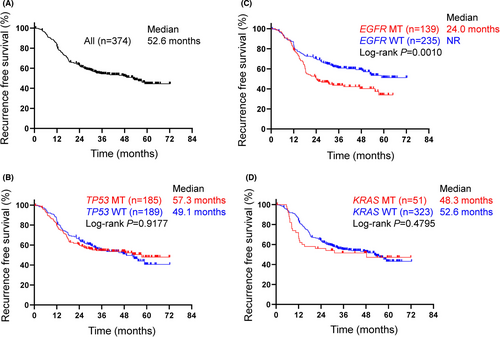
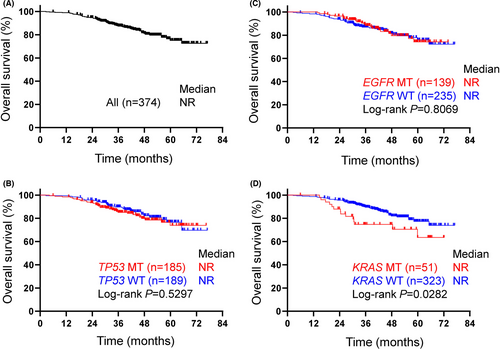
No difference in RFS or OS was observed between patients with or those without the TP53 mutation (Figures 3B and 4B). The median RFS of patients with common EGFR mutations was significantly shorter than that of patients with wild-type EGFR (24.0 mo vs not reached, P = .0010, log-rank) (Figures 2 and 3C). Conversely, no difference in OS was observed between the patients with an EGFR mutation and those with wild-type EGFR (Figure 4C). When focusing on the major EGFR mutations, Ex19Del and L858R, the RFS period of the patients with Ex19Del (vs wild-type, P = .0056) or L858R (P = .0275) was shorter than that of patients with wild-type EGFR (Figure S1). No difference in the OS period was observed between patients with an Ex19Del mutation and those with L858R. The EGFR mutation was likely to be a predictive factor for recurrence. In addition, a significant difference of RFS period between patients with EGFR-mutant and wild-type genotype was observed in stage III but not stage II patients, although the reason remains unclear (Figures S2A,B).
In our EGFR mutation-positive population, there were no differences in clinical outcomes of platinum-based chemotherapy between TP53-positive and TP53-negative patients (Figure S3). The median RFS of the patients with a KRAS mutation was marginally shorter than that of those with wild-type KRAS, but the difference was not significant (48.3 vs 52.6 mo, log-rank P = .4795) (Figure 3D). The median OS of the patients with KRAS mutations was significantly shorter than that of patients with wild-type KRAS (Figure 4D). KRAS mutation was likely to be a poor prognostic factor.
3.2 The relationship between EGFR mutations and prognosis
The relationship between the EGFR mutation status and clinicopathological factors was investigated using the Fisher exact test (Table 1). The EGFR mutation was frequently detected in female patients (P < .0001), as reported previously.13, 14 Univariate analysis showed that the EGFR mutation, female sex, and advanced stage were associated with poor postoperative recurrence in patients with NSCLC (Figure 5A). Multivariate analysis showed that advanced stage and EGFR mutation status were independent risk factors for postoperative recurrence (HR, 1.78; 95% CI, 1.30-2.44; P = .0003 for stage; and HR, 1.57; 95% CI, 1.15-2.16; P = .0050 for EGFR mutation status) (Figure 5B). Disease stage (P = .0002) and EGFR mutation (P = .0019) were retained after backwards elimination (data not shown). These results suggest that stage II-III Ns-NSCLC patients with EGFR mutations may have a shorter RFS.
| Characteristics | EGFR mutation (n = 139) | EGFR wild-type (n = 235) | P |
|---|---|---|---|
| Treatment | |||
| Vnr/Cis | 78 (56.1) | 115 (48.9) | .1994 |
| Pem/Cis | 61 (43.9) | 120 (51.1) | |
| Sex | |||
| Male | 55 (39.6) | 170 (72.3) | < .0001 |
| Female | 84 (60.4) | 65 (27.7) | |
| Age | |||
| ≥70 y | 33 (23.7) | 44 (18.7) | .2898 |
| <70 y | 106 (76.3) | 191 (81.3) | |
| PS | |||
| 0 | 108 (77.7) | 171 (72.8) | .3263 |
| 1 | 31 (22.3) | 64 (27.2) | |
| Stage | |||
| IIA/IIB | 55 (39.6) | 109 (46.4) | .2356 |
| IIIA | 84 (60.4) | 126 (53.6) | |
Note
- Univariate analysis of clinicopathological factors for patients with or without EGFR mutation. Significantly more EGFR mutations were found in female patients by Fisher exact test.

4 DISCUSSION
Our results showed that stage II-III Ns-NSCLC patients with EGFR-sensitizing mutations had a shorter median RFS, but not a shorter OS, than those without such mutations after the treatment with adjuvant chemotherapy with platinum doublet chemotherapy (Pem/Cis or Vnr/Cis). This difference in RFS may be due to either prognostic factors or the poor effect of chemotherapy in patients with EGFR mutations. The prognostic values for EGFR mutations have been previously investigated in retrospective studies.5-7
EGFR-TKI treatment influences OS, especially in patients with advanced NSCLC. However, whether or not platinum-based adjuvant chemotherapy improves the prognosis of patients with EGFR-mutated NSCLC has been controversial.5, 6 In previous studies, the presence of EGFR mutations was confounded by favorable prognostic factors, such as a female sex and a non-smoking status, and the number of cases, even if adjusted in a multivariate analysis, might not be sufficient to eliminate the effect of EGFR mutations. In the present study, patients with EGFR mutations were also treated with EGFR-TKIs as follow-up therapy. It is likely that this follow-up therapy influenced OS in our study. Conversely, patients with EGFR mutations who were treated with platinum doublet therapy clearly demonstrated a significantly shorter RFS in our prospective study. Whether the EGFR mutation status is prognostic or predictive of a response to platinum doublet therapy remains unclear. In solid cancers including NSCLC, oncogene alterations such as RAS, HER2, and MET were poorly prognostic.15-17 The KRAS mutation is known to be a poor prognostic factor.18, 19 In our cohort, patients with the KRAS mutation had a shorter RFS (P = .4795) and OS (P = .0282). An oncogene HER2 mutation in NSCLC also reportedly predicts a poor prognosis.20 Based on the hypothesis that the oncogenic potential of EGFR mutations is associated with a poor prognosis, the potential difference in driver oncogenes between Ex19Del and L858R may result in a difference in RFS.
Biologically, ligand binding promotes EGFR dimerization, which determines a series of structural rearrangements that are conveyed to the cytoplasmic domain and allow the formation of asymmetric dimers between 2 juxtaposed catalytic domains. The EGFR mutation is constitutively active without requiring ligand stimulation.21 EGFR-sensitizing mutations have been shown to exert tumorigenicity in transgenic mice.11 Dimerization is required for the activation of the cellular signaling of L858R, but not Ex19Del.22 Ex19Del exerts stronger kinase activity, tumorigenicity, and a higher sensitivity to EGFR-TKI than L858R.23, 24 Ex19Del is therefore thought to act as a more potent driver oncogene than L858R.24 It has been argued that the biological differences between Ex19Del and L858R may be responsible for the different effects of EGFR-TKIs.25-27 Lee and colleagues28 reported a meta-analysis for NSCLC patients treated with chemotherapy and demonstrated a shorter progression-free survival (PFS) among patients with Ex19Del, compared with those with L858R. In our cohort, the RFS period of the patients with both Ex19Del and L858R was significantly shorter than that of patients with wild-type EGFR, but a long-tail of the curve was observed for L858R but not Ex19Del (Figure S1).
It has been reported that the concurrent TP53 mutation was associated with unfavorable efficacy to EGFR-TKI in patients with EGFR-mutated NSCLC.29 In adjuvant platinum-based chemotherapy, the TP53 mutation had no prognostic effect on patients with NSCLC from adjuvant cisplatin-based therapy randomized trials.30, 31 Shepherd and colleagues32 could identify no prognostic effect of co-mutation of TP53 and EGFR mutation on NSCLC patients with adjuvant chemotherapy. In our EGFR mutation-positive population, there were no differences in clinical outcomes of platinum-based chemotherapy between TP53-positive and TP53-negative patients (Figure S3).
For adjuvant chemotherapy with osimertinib in the ADAURA study, the 2-y RFS in the placebo arm was 44%, similar to the results of our analysis.5 For gefitinib as adjuvant chemotherapy (CTONG 1104), the 3-y DFS in the cisplatin plus vinorelbine arm was 32.5%, which was slightly worse than that in our study.7 These results support the hypothesis that NSCLC patients with EGFR mutation have a poor prognosis.
The limitations of this study were as follows: (a) no analysis of RFS and OS was performed for patients with uncommon EGFR mutations because of the limited number of uncommon mutations; and (b) the effect of fusion genes could not be analyzed. Despite these limitations, we believe that the evidence from this prospective clinical trial provides some reliable data.
In conclusion, EGFR mutation-positive NSCLC in patients with stage II-III disease is either of poor prognostic or predictive impact on a poor response to postoperative chemotherapy with platinum doublets. This result supports the mandatory molecular diagnosis of early-stage NSCLC for EGFR-TKI therapy and precision oncology. Wu and colleagues5 reported in patients with stage IB to IIIA EGFR mutation-positive NSCLC, DFS was significantly longer among those who received osimertinib than among those who received placebo. The present study supports the importance of adjuvant for the 3rd generation tyrosine kinase inhibitor rather than platinum-based chemotherapy.
ACKNOWLEDGMENTS
This research was supported by University Grants for Fundamental Research of Kindai University. We thank the participating patients and their families as well as all the site investigators and operations staff. We are grateful to data managers and other support staff of the West Japan Oncology Group, especially Mr. Sawa, Ms. Tanaka, Dr. Nakamura, and Dr. Takeda. The authors also thank Mr. Mine (Center for Instrumental Analyses Central Research Facilities, Kindai University Faculty of Medicine) and Ms. Kitano (Department of Genome Biology, Kindai University Faculty of Medicine) for technical assistance provided during the study.
DISCLOSURE
Toshiaki Takahashi reports grants and personal fees from AstraZeneca, Chugai Pharmaceutical, Eli Lilly, Ono Pharmaceutical, MSD, Pfizer, Nippon Boehringer Ingelheim, a grant from Amgen, and a personal fee from Roche Diagnostics, outside of the submitted work. Kazuko Sakai received personal fees from AstraZeneca, Bio-Rad Laboratories, Chugai Pharmaceutical, Roche Diagnostics, Hitachi, outside of the submitted work. Hirotsugu Kenmotsu received grants and personal fees from Chugai Pharmaceutical, Novartis Pharma, Daiichi Sankyo, AstraZeneca, and personal fees from Ono Pharmaceutical, Boehringer Ingelheim, Eli Lilly, Kyowa Kirin, Bristol-Myers Squibb, MSD, Pfizer, Taiho Pharmaceutical, outside of the submitted work. Kiyotaka Yoh received grants and personal fees from AstraZeneca, Eli Lilly, Daiichi Sankyo, Taiho Pharmaceutical, grants from Pfizer, AbbVie, Bayer, Takeda Pharmaceutical, MSD, and personal fees from Bristol-Myers Squibb, Chugai Pharmaceutical, Janssen, Novartis, Kyowa Kirin, Boehringer Ingelheim, outside of the submitted work. Haruko Daga received personal fees from AstraZeneca, Chugai Pharmaceutical, Eli Lilly Japan, MSD, Ono Pharmaceutical, Taiho Pharmaceutical, outside the submitted work. Hidetoshi Hayashi received grants and personal fees from AstraZeneca, Bristol-Myers Squibb, Eli Lilly, MSD, Ono Pharmaceutical, Pfizer, Nippon Boehringer Ingelheim, Novartis Pharma, Merck Biopharma, Taiho Pharmaceutical, grants from AbbVie, AC Medical, Astellas Pharma, Daiichi Sankyo, Eisai, EPS Associates, GlaxoSmithKline, Japan Clinical Research Operations, Kyowa Kirin, Otsuka Pharmaceutical, Parexel International, PPD-SNBL, Quintiles Transnational Japan, Takeda Pharmaceutical, Yakult Honsha, and a personal fee from Chugai Pharmaceutical, outside of the submitted work. Yuichi Takiguchi received grants and personal fees from Eli Lilly, Kyowa Kirin. Akimasa Sekine received personal fees from Eli Lilly, Ono Pharmaceutical, Chugai Pharmaceutical, AstraZeneca, Pfizer, outside the submitted work. Yuki Sato received personal fees from Novartis Pharma, Chugai Pharmaceutical, AstraZeneca, Pfizer, outside the submitted work. Hiroaki Akamatsu received grants and personal fees from Chugai Pharmaceutical, MSD, and personal fees from AstraZeneca, Nippon Boehringer Ingelheim, Bristol-Myers Squibb, Eli Lilly, Novartis Pharma, Ono Pharmaceutical, Taiho Pharmaceutical, outside of the submitted work. Takashi Seto received grants and personal fees from Chugai Pharmaceutical, Daiichi Sankyo, Eli Lilly, MSD, Novartis Pharma, Pfizer, Takeda Pharmaceutical, grants from AbbVie, Kissei Pharmaceutical, Merck Biopharma, and personal fees from AstraZeneca, Bristol-Myers Squibb, Covidien Japan, Kyowa Kirin, Mochida Pharmaceutical, Nippon Boehringer Ingelheim, Ono Pharmaceutical, Taiho Pharmaceutical, Thermo Fisher Scientific, outside of the submitted work, and is an employee of Precision Medicine Asia. Kenji Sugio received grants and personal fees from AstraZeneca, Chugai Pharmaceutical, Eli Lilly, Taiho Pharmaceutical, Nippon Boehringer Ingelheim, Merck Sharp & Dohme Oncology, outside of the submitted work. Makoto Nishio received grants and personal fees from Ono Pharmaceutical, Bristol-Myers Squibb, Pfizer, Chugai Pharmaceutical, Eli Lilly, Taiho Pharmaceutical, AstraZeneca, MSD, Novartis, Daiichi Sankyo, Merck Serono, Takeda Pharmaceutical, Pfizer, Janssen, and personal fees from Boehringer Ingelheim, Nippon Kayaku, and consulting fees from AbbVie, Teijin Pharma, Ono Pharmaceutical, Chugai Pharmaceutical, Taiho Pharmaceutical, Bristol-Myers Squibb, Takeda Pharmaceutical, Pfizer, Daiichi Sankyo, Eli Lilly, AstraZeneca, MSD outside the submitted work. Nobuyuki Yamamoto received grants and personal fees from AstraZeneca, Nippon Boehringer Ingelheim, Chugai Pharmaceutical, Eli Lilly, MSD, Ono Pharmaceutical, Pfizer, grants from Astellas Pharma, A2 Healthcare Corporation, Bristol-Myers Squibb, CMIC Shift Zero, Daiichi Sankyo, IQVIA services Japan, PPD-SNBL, Takeda Pharmaceutical, Taiho Pharmaceutical, and a personal fee from Novartis Pharma, outside the submitted work. Kazuto Nishio received grants and personal fees from Eli Lilly, Nippon Boehringer Ingelheim, grants from Ignyta, Korea Otsuka Pharmaceutical, Thoracic Oncology Research Group, North East Japan Study Group, and personal fees from Chugai Pharmaceutical, Eisai, Pfizer, Novartis Pharma, MSD, Ono Pharmaceutical, Bristol-Myers Squibb, SymBio Pharmaceuticals, Life Technologies Japan, Solasia Pharma, Yakult Honsha, Roche Diagnostics, AstraZeneca, Otsuka Pharmaceutical, Sanofi, Guardant Health, Amgen, outside of the submitted work. Masahiro Tsuboi received grants and personal fees from AstraZeneca, Ono Pharmaceutical, Bristol-Myers Squibb, Eli Lilly, MSD, grants from Nippon Boehringer Ingelheim, and personal fees from Novartis Pharma, Johnson and Johnson, Chugai Pharmaceutical, Teijin Pharma, Taiho Pharmaceutical, Medtronic Japan, outside the submitted work. The remaining authors have declared no conflicts of interest.
ETHICAL APPROVAL
This study was conducted in compliance with the Helsinki Declaration and the Ethical Guidelines for Medical and Health Research Involving Human Subjects by the Japanese government. The study was also approved by the ethics committee of each participating institute.
Open Research
DATA AVAILABILITY STATEMENT
Data generated or analyzed during this study are available from the corresponding author on reasonable request.




