Long non-coding RNAs and prostate cancer
Funding Information
This work was supported by grants of P-DIRECT and P-CREATE from Ministry of Education, Culture, Sports, Science and Technology, Japan (SI); by grants (K.T. and S.I.) from JSPS (numbers 15K15581, 15K15353), Japan; by a grant from the Program for Promotion of Fundamental Studies in Health Sciences from the National Institute of Biomedical Innovation, Japan (S.I.); by Grants-in-Aid (S.I.) from the Ministry of Health, Labor and Welfare, Japan; and grants from the Terumo Foundation for Life Sciences and Arts (K.T.) and the NOVARTIS Foundation for the Promotion of Science, Japan (K.T.).
Abstract
Long non-coding RNAs (lncRNAs) are RNA transcripts larger than 200 nucleotides that do not code for proteins the aberrant expression of which has been documented in various types of cancer, including prostate cancer. Lack of appropriate sensitive and specific biomarkers for prostate cancer has led to overdiagnosis and overtreatment, making lncRNAs promising novel biomarkers as well as therapeutic targets for the disease. The present review attempts to summarize the current knowledge of lncRNA expression patterns and mechanisms in prostate cancer, which contribute to carcinogenesis. In particular, we focused on lncRNAs regulated by androgen receptor and expressed in castration-resistant prostate cancer.
Non-coding RNAs, which are RNA transcripts that do not code for proteins, can be divided into two major groups: small ncRNAs between 18 and 200 nt in length, and lncRNAs, which are larger than 200 nt. miRNAs are evolutionally conserved and single-stranded small non-protein coding transcripts of approximately 18–22 nt that post-transcriptionally regulate gene expression.1 Aberrant expressions of miRNAs have been well documented in most types of cancer.2 Unlike miRNAs, lncRNAs are able to fold into secondary and tertiary structures by which they carry out their function3 Several studies have shown the importance of lncRNAs as modulators of key cellular processes not only in normal physiology4, 5 but also in diseases such as cancer, including prostate cancer.6-8 It is assumed that many of these transcripts could serve as future cancer biomarkers.9
Prostate cancer is one of the leading causes of cancer morbidity and mortality in developed countries. It is currently diagnosed based on the measurement of serum PSA, also known as KLK3, and DRE. Treatment of localized prostate cancer takes into account clinicopathological factors including Gleason score, initial PSA level, patient's age and clinical tumor stage.10 Active surveillance may be best for patients with low-risk disease (i.e. Gleason score of 6 or less), whereas men with high-risk disease (i.e. Gleason score >7, PSA levels >20 ng/mL and clinical tumor stage >pT2c, i.e. tumor involving both prostate lobes) will instead benefit from radical prostatectomy or radiotherapy.11 AR and its downstream signaling are fundamental for the development and progression of both localized and advanced metastatic prostate cancer.12 Therefore, nowadays, the recommended therapy for locally advanced prostate cancer consists of long-term ADT in combination with radiotherapy. ADT decreases circulating testosterone levels to a very low amount (<50 ng/mL), a condition called chemical castration of men. However, some tumors will become hormone refractory following ADT, featured by increasing PSA levels in blood and upregulation of the AR in cancer cells.13, 14 Over a period of 12–36 months, a disease state called CRPC evolves in many patients. The ineffectiveness of conventional ADT in these CRPC is a result of activation of AR and its downstream pathways.14 Past studies have revealed that elevated AR expression,15 enhanced AR activity16 and development of AR variants17 are observed in the progression of CRPC. Therefore, identification of AR downstream signals and new molecular mechanisms for AR activation are important to improve the treatment of advanced prostate cancer.
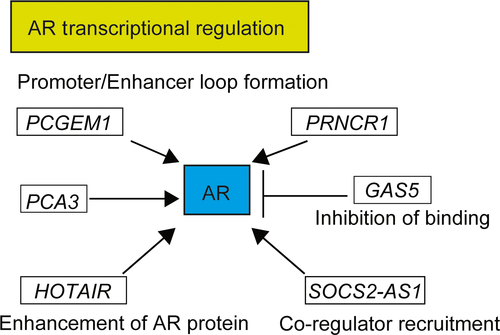
LncRNAs are aberrantly expressed in a variety of human diseases, contributing to pathogenesis or maintaining diseased conditions.8 Aberrantly expressed lncRNAs can be indicative of certain stages of cancer progression, and may predict early progression or efficiently sustain tumor-related signaling pathways.8 Thus, lncRNAs have the potential to be applicable for the diagnosis of prostate cancer, as well as being potential criteria in the choice of therapy and potential new therapeutic targets of CRPC. The present review attempts to summarize the current knowledge of lncRNA expression patterns in prostate cancer and the mechanisms that contribute to prostate carcinogenesis focusing on AR-regulated lncRNAs and lncRNAs expressed in CRPC (Fig. 1).
LncRNAs and Prostate Cancer
LncRNAs as biomarkers in prostate cancer
PCA3 is one of the most specific prostate cancer biomarkers, which was originally discovered in 1999 by differential display analysis of prostate tissues and cell lines.18 Its expression was shown to be 60–100-fold higher in more than 95% of prostate tumors compared to adjacent non-neoplastic tissues, and is undetectable in other tumor types. PCA3 knockdown inhibits AR signaling, cell growth and viability, suggesting that overexpression of PCA3 may modulate AR signaling in tumor cells. Knockdown of PCA3 leads to partial upregulation of epithelial markers such as E-cadherin, claudin-3 and cytokeratin-18, and downregulation of the mesenchymal marker vimentin.19 PCA3 also regulates the expression of important cancer-related genes involved in apoptosis, angiogenesis, signal transduction, cell adhesion and mitogen-activated kinase kinase 1.19 In addition, a working model of PCA3 has been proposed, in which PCA3 acts as a dominant-negative oncogene that downregulates the unrecognized tumor suppressor Prune Homolog 2 (PRUNE2), a human homolog of the Drosophila prune gene, through a process that involves RNA editing by the formation of PRUNE2/PCA3 double-stranded RNA.20 Combination of urinary PCA3 and fusion gene TMPRSS2-ERG can increase specificity in prostate cancer diagnosis compared with serum PSA, and has the potential to substantially reduce unnecessary prostate biopsies. (Functions of lncRNAs in prostate cancer and references are summarized in Table 1 and Fig. 2).
| Expression in PCa | LncRNAs | Role | Implications in PCa | References |
|---|---|---|---|---|
| ↑ | PCA3 | Biomarker | Enhances AR signaling, cell growth and viability. Regulates the expression of important cancer-related genes | 18-20 |
| ↑ | SChLAP1 | Overexpression is associated with risk of biochemical recurrence, clinical progression, metastasis and PCa-specific mortality | 21-24 | |
| ↑ | SPRY4-IT1 | siRNA knockdown inhibits cell proliferation and invasion, and increases apoptosis | 25 | |
| ↑ | MALAT1 | Overexpression is associated with indicators of poor prognosis. Binds to EZH2 to enhance migration and invasion | 26-31 | |
| ↑ | TRPM2-AS | Transcribed from the antisense strand of TRPM2, which is activated with TMPR2-AS knockdown. Overexpression associated with poor prognosis | 32, 33 | |
| ↑ | NEAT1 | Expression associated with PCa cell progression. Alters the epigenetic status of target genes to drive oncogenic growth | 34 | |
| ↑ | PCGEM1 | AR-related | Promotes cell proliferation by regulating c-Myc | 35, 37, 46, 47 |
| ↑ | PCAT1 | Upregulated in high-grade localized and metastatic PCa. Promotes cell proliferation by regulating c-Myc. Represses BRCA2 | 38, 39 | |
| ↑ | PCAT6 | Predictive of tumor progression by AR signaling. Overexpressed in primary and metastatic PCa. siRNA-mediated knockdown reduces cell growth | 8, 40, 41 | |
| ↑ | PCAT7 | |||
| ↑ | PCAT18 | Predictive of tumor progression by AR signaling. Metastatic PCa specific. Induced by AR. siRNA-mediated knockdown reduces cell growth | 8, 42 | |
| ↑ | PRNCR1 | Associated with prostate cancer susceptibility. siRNA knockdown attenuates cell viability and AR activity. Could be involved in prostate carcinogenesis through AR | 44, 45 | |
| ↑ | CTBP1-AS | Transcribed from the antisense strand of CTBP1. Promotes castration-resistant prostate tumor growth by regulating epigenetically cancer-associated genes | 46, 49 | |
| ↑ | HOTAIR | Repressed by androgen and upregulated in CRPC after deprivation therapies. Binds to AR to prevent its degradation. Overexpression increases cell growth and invasion | 58 | |
| ↑ | SOCS2-AS1 | Transcribed from the antisense strand of SOCS2. Promotes cell proliferation, migration and anti-apoptosis. | 59 | |
| ↑ | POTEF-AS1 | Transcribed from the antisense strand of POTEF. Promotes cell proliferation and anti-apoptosis | 60 | |
| ↓ | GAS5 | Tumor suppressor | Represses AR action and promotes apoptosis. Downregulated in CRPC. Reciprocal regulation of GAS5 levels and mTOR inhibitor action | 62-64 |
| ↓ | H19 | Upregulation of H19 represses cell migration. H19-derived miR-675 targets TGFβ1 to repress cell migration | 69 | |
| ↓ | PCAT29 | First AR-repressed lncRNA that functions as a tumor suppressor. Low PCAT29 expression correlated with poor prognostic outcomes. Overexpression suppresses cell growth and metastasis | 43 |
- AR, androgen receptor; BRCA2, breast cancer susceptibility gene 2; CRPC, castration-resistant prostate cancer; CTBP1, C-terminal binding protein 1; EZH2, enhancer of zeste homolog 2; GAS5, growth arrest-specific 5; lncRNAs, long non-coding RNAs; mTOR, mammalian target of rapamycin; PCa, prostate cancer; POTEF, prostate, ovary, testis expressed protein family member-F gene; SOCS2, suppressor of cytokine signaling 2; TGF-β1, transforming growth factor beta 1; TRPM2-AS, transient receptor potential cation channel, subfamily M, member 2-antisense transcript.

Second chromosome locus-associated with prostate-1 (SChLAP1) is a lncRNA highly expressed in 25% of prostate cancer.21 Its expression is more frequent in CRPC and it is significantly associated with risk of biochemical recurrence, clinical progression, metastasis and prostate cancer-specific mortality.21, 22 SChLAP1 interacts with Switch-Sucrose Non-Fermentable (SWI/SNF) complex for chromatin remodeling, counteracting the tumor-suppressor effects of SWI/SNF.21 Analysis of SChLAP1 expression by ISH showed that this lncRNA independently predicts biochemical recurrence after radical prostatectomy.23 Furthermore, SChLAP1 expression also correlated with prostate cancer lethal progression, which makes this lncRNA a useful tissue-based biomarker for identifying PCa patients at higher risk of CRPC progression.24
SPRY4 intronic transcript 1 (SPRY4-IT1) is one of the lncRNAs highly upregulated in PC3 cells and in patient samples compared to non-malignant prostate epithelial cells and matched normal prostate tissues.25 siRNA knockdown of SPRY4-IT1 inhibited PC3 cellular proliferation and invasion, and increased apoptosis.25 SPRY4-IT1 was easily detected in all prostate cancer samples with different Gleason scores (6–10) in an RNA chromogenic ISH assay.25 Prostate cancer specificity and easy detection with standard clinical staining procedures of tissue samples makes this lncRNA a useful candidate as a diagnostic biomarker.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a lncRNA originally found to be overexpressed in non-small-cell lung cancer tissues with a high risk of metastasis.26 Recent studies showed that MALAT1 was also overexpressed in other human cancers, including breast, pancreas, colon, prostate, and liver cancers.27, 28 In prostate cancer, MALAT1 overexpression was associated with indicators of poor prognosis such as high Gleason score, higher tumor-node-metastasis stage and serum PSA >20 ng/mL, and its expression was significantly higher in CRPC than in hormone-sensitive prostate cancer.29 In a study comparing the expression of MALAT1 in urinary samples of biopsy-positive and biopsy-negative prostate cancer patients, this lncRNA was significantly higher in biopsy-positive samples,30 suggesting that urinary MALAT1 may be a promising diagnostic biomarker. Furthermore, using EZH2 antibody-based RNA immunoprecipitation coupled with high throughput sequencing analysis, it was demonstrated that MALAT1 binds to EZH2.31 It was indicated that MALAT1 plays a crucial role in EZH2-enhanced migration and invasion in CRPC cell lines, and a positive correlation between MALAT1 and EZH2 has been documented.31
Transient receptor potential cation channel, subfamily M, member 2-antisense transcript (TRPM2-AS) is an antisense lncRNA transcribed from the opposite strand of the TRPM2 gene that is overexpressed in prostate cancer.32 High TRPM2-AS was associated with poor prognosis and in vitro TRPM2-AS knockdown led to prostate cancer cell apoptosis and activation of the TRPM2 gene. Microarray analysis was carried out using PC3 cells which were transfected with an siRNA ablating TRPM2-AS RNA or control siRNA to analyze the mechanisms by which TRPM2-AS maintains cell survival in prostate cancer cells.33 Results showed that TRPM2-AS coordinates the expression of a large number of genes involved in controlling survival, unfolded protein response and cell cycle in prostate cancer cells. Moreover, targets of existing drugs and treatments were found to be consistently regulated by TRPM2-AS knockdown. Thus, the essential role of TRPM2-AS as a key regulator of survival in prostate cancer makes this lncRNA a suitable therapeutic target for further clinical studies.
Estrogen receptor alpha (ERα) is expressed in prostate cancer, independent of AR status and, in CRPC, ERα signaling constitutes an effective mechanism to bypass the androgen/AR axis. In a study combining ChIP- and RNA-seq data in prostate cancer cells, an ERα-specific non-coding transcriptome signature was identified, where the lncRNA, nuclear enriched abundant 1 (NEAT1), was the most significantly overexpressed lncRNA in prostate cancer.34 NEAT1 expression was associated with prostate cancer progression, and prostate cancer cells expressing high levels of this lncRNA were resistant to androgen deprivation or AR antagonists. NEAT1 is recruited to the promoter regions of target genes and alters their epigenetic status to favor transcription and drive oncogenic growth. NEAT1 levels increase after long-term androgen-deprivation therapies, and its expression is significantly higher in CRPC compared to hormone-sensitive prostate cancer, which indicates that ERα-NEAT1 interaction may promote castration resistance. Elevated NEAT1 levels are associated with early biochemical recurrence and metastatic spread, making this lncRNA both a potential therapeutic target and a reliable prognostic biomarker.
LncRNAs related with AR signaling
Prostate cancer gene expression marker 1 (PCGEM1) and prostate cancer-associated ncRNA transcript 1 (PCAT1) are highly prostate-specific lncRNAs and considered attractive biomarkers. PCGEM1 was one of the oncogenic lncRNAs identified in prostate cancer and regulated by androgen.35 PCGEM1 is expressed in at least half of prostate tumors and functional analysis revealed oncogenic properties such as promotion of cell proliferation and colony formation.36 It was demonstrated that PCGEM1 promotes cancer cell proliferation by regulating c-Myc.37 PCAT1 is a lncRNA which is expressed highly specifically in prostate and upregulated in high-grade localized (Gleason score >7) and metastatic prostate cancer. PCAT1 induces cell proliferation in vitro and has repressive effects on a broad range of genes, including the tumor suppressor gene, breast cancer susceptibility gene 2 (BRCA2).38 PCAT1 also promotes cell proliferation by interaction with c-Myc by acting as a decoy for c-Myc-targeting miRNAs.39 Other PCAT-family members, PCAT6, PCAT 7, and PCAT 18 were predictive of tumor progression by modulation of AR signaling.8, 40 Expressions of PCAT6 and PCAT7 were higher in primary and metastatic prostate cancer, and siRNA-mediated knockdown of either of them reduced cell growth and soft agar colony formation in both androgen-dependent LNCaP and -independent LNCaP sublines.41 PCAT18 is highly prostate-specific and expressed especially in metastatic prostate cancer tissues. Expression level of PCAT18 is regulated by AR signaling.42 siRNA-mediated knockdown of PCAT18 also reduced cell growth. PCAT29 was identified as an AR-repressed lncRNA that functions as a tumor suppressor.43 PCAT29 was suppressed by androgen and upregulated by castration in a prostate cancer xenograft model. PCAT29 knockdown significantly increased proliferation and migration of prostate cancer cells, whereas PCAT29 overexpression conferred the opposite effect. In prostate cancer patient specimens, low PCAT29 expression correlated with poor prognostic outcomes, suggesting that decrease of this lncRNA may identify a subset of patients at higher risk for disease recurrence.
Prostate cancer noncoding RNA-1 (PRNCR1) was initially identified as a novel lncRNA transcribed from 8q24, the genomic locus most significantly associated with prostate cancer susceptibility found in a genome-wide association study.44 Knockdown of PRNCR1 by siRNA attenuated prostate cancer cell viability and AR transactivation activity, indicating that PRNCR1 could be involved in prostate carcinogenesis possibly through AR. In another study, PRNCR1 and PCGEM1 were found to successively bind to AR and strongly enhance AR-mediated gene activation programs and proliferation in prostate cancer cells.45 Later, it was shown that PCGEM1 and PRNCR1 do not interact with AR and that neither gene was a prognostic factor for prostate cancer.46 A study using AR+/androgen-dependent prostate cancer xenograft models revealed that PRNCR1 was scarcely expressed and that PCGEM1 expression was lower in metastatic prostate cancer compared to primary tumors.47 Thus, the roles of these lncRNAs in prostate cancer are still being debated, and further studies are necessary to elucidate the relation of PCGEM1 and AR, and its clinical significance.
Our study analyzing a global AR transcriptional network by mapping genome-wide androgen-regulated TTS using CAGE and ARBS using ChIP-seq has revealed comprehensive AR-regulated transcripts from intergenic or AS regions of genes in prostate cancer cells.48 Based on this study, a lncRNA located at the AS region of the C-Terminal Binding Protein 1 (CTBP1) gene, CTBP1-AS, was found to promote tumor growth of both hormone-sensitive prostate cancer and CRPC models by regulating cancer-associated genes epigenetically.49 These studies revealed the importance of androgen-regulated lncRNAs in prostate cancer progression.
HOX Transcript Antisense RNA (HOTAIR) is a well-known lncRNA found to play important roles in several tumors.50-57 In prostate cancer, HOTAIR was reported as a lncRNA repressed by androgen.58 It was markedly upregulated in CRPC after androgen deprivation therapies. HOTAIR was found to bind to AR protein to block its interaction with E3 ubiquitin ligase Mouse double minute 2 homolog (MDM2), preventing AR ubiquitination and degradation. HOTAIR overexpression increased prostate cancer cell growth and invasion. Consequently, HOTAIR expression was sufficient to induce androgen-independent AR activation and drive an AR-mediated transcriptional program in the absence of androgen. HOTAIR may drive androgen-independent AR activity and CRPC progression, being a potential therapeutic target.
Our comprehensive analysis of AR-regulated lncRNAs in two prostate cancer cell lines and CRPC model cells by directional RNA-seq analysis identified an androgen-regulated lncRNA, suppressor of cytokine signaling 2-antisense transcript 1 (SOCS2-AS1), located at the antisense strand of SOCS2. Expressions of SOCS2-AS1 and SOCS2 expression were androgen-dependent and their expression was suppressed by AR knockdown as well as by an anti-androgen, bicalutamide. SOCS2-AS1, which is highly expressed in CRPC model cells, promoted castration-resistant and androgen-dependent cell growth and inhibited apoptosis. Furthermore, SOCS2-AS1 promoted androgen signaling by modulating the epigenetic control for AR target genes including Tumor Necrosis Factor Superfamily Member 10 (TNFSF10), suggesting that SOCS2-AS1 plays an important role in the development of CRPC by repressing apoptosis.59 (Fig. 3.)
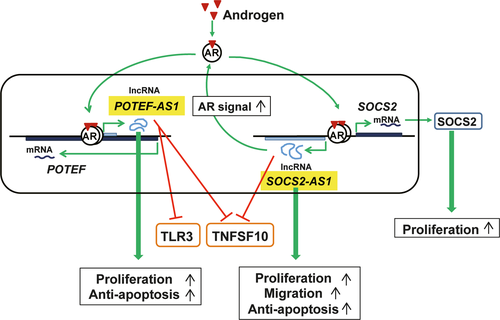
We recently identified another AR-dependent lncRNA transcribed from the AS strand of prostate, ovary, testis expressed protein family member-F gene (POTEF), which was named POTEF-Antisense transcript-1 (POTEF-AS1).60 POTEF is one of the proteins encoded by the POTE family genes, which are preferentially expressed in prostate, ovary, testis and placenta.61 POTEF-AS1 promoted cell growth, inhibited apoptosis and repressed genes related to TLR signaling and apoptosis pathways in LNCaP cells, suggesting that POTEF-AS1 would play a key role in the progression of prostate cancer by repressing TLR signaling.60 (Fig. 3.)
LncRNAs as tumor suppressors
Growth arrest-specific 5 (GAS5) is a tumor suppressive lncRNA that promotes apoptosis and represses AR action by sequestering androgen/AR complex and preventing binding of the complex to target genes.62 Its expression level was shown to decline when prostate cancer cells acquire castrate resistance.63 Recently, regulation of GAS5 expression by mTOR inhibitor was found in prostate cancer cells. Several mTOR inhibitors increased cellular GAS5 expression levels and inhibited cell growth in AR-positive LNCaP and 22Rv1 cells, whereas silencing of GAS5 in these cells decreased sensitivity to mTOR inhibitors.64 In early-stage prostate cancer, mTOR inhibitors may be used to increase GAS5 levels to enhance cellular apoptosis.
Genomic imprinting is a form of epigenetic gene regulation that results in expression from a single allele in a parent-of-origin-dependent manner. One of the most extensively studied imprinting-associated lncRNAs in cancer is Long Intergenic Non-Protein Coding RNA 8 (H19).65 H19 is located in an imprinted region of chromosome 11 near the insulin-like growth factor 2 (IGF2) gene, the former expressed only from the maternally inherited chromosome, whereas the latter is expressed only from the paternally inherited chromosome.66 H19 was found to be a lncRNA which plays a critical role in choriocarcinoma67 and bladder cancer.68 However, in metastatic prostate cancer, H19 has a tumor suppressive role by repressing the effects of transforming growth factor beta 1 (TGFβ1). H19 and H19-derived miR-675 were both significantly downregulated in metastatic prostate cancer cells compared to non-metastatic prostate cancer cells.69 Upregulating H19 increased miR-675 levels and repressed cell migration. In addition, miR-675 repressed TGFβ1 translation by directly binding to the 3′UTR.
Clinical Use of lncRNAs and Limitations
LncRNAs still remain uncharacterized RNA molecules as a result of their low expression levels, poor conservation, and high tissue/cell specificity.70 In prostate cancer diagnosis, the main priority is identifying novel biomarkers to reliably distinguish between low-risk and high-risk patients who need definitive treatment.71 Although the majority of studies have focused on the intracellular roles of lncRNAs, there is an increasing interest in the potential roles of extracellular circulating lncRNAs. Urinary PCA3 is now widely used as a biomarker for detection of cancer and has been approved by the United States Food and Drug Administration.72 Measuring PCA3 was shown to be superior to serum PSA test and digital rectal examination in the sensitivity and specificity for prostate cancer diagnosis.73, 74 However, its utility as a first-line test or to detect high-grade prostate cancer remains under investigation.75 Therefore, other methods for prostate cancer diagnosis using lncRNAs have been developed, such as MALAT1-derived (MD) mini-RNA (MD-mini-RNA).76 Plasma MD-mini-RNA was found to improve diagnostic accuracy for predicting the results of prostate biopsy in patients with high PSA levels (>4 ng/mL). These circulating lncRNAs have shown promise as non-invasive biomarkers across a wide spectrum of diseases and altered physiological states.77, 78 Release of cellular RNA species into circulation may reflect disease-specific tissue injury and/or remodeling, or potential intercellular signaling. However, the biological functions and the mechanisms of circulating lncRNAs have not been elucidated. Uniform normalization protocol of sample collection, lncRNAs extraction, endogenous control selection, quality assessment, and quantitative data analysis has not been established. To adopt circulating lncRNA into clinical practice, the following aspects should be investigated: normalization of sample preparation such as temperature, volume and reagents used to extract and store lncRNAs, selection of reliable endogenous controls, method to assess the quality of lncRNA and reduction of bias by increasing the size of the research cohort.
FANTOM Projects to Investigate the Functional Role of lncRNAs
Recent advances in transcriptomics have improved coverage as well as detection accuracy of profiled RNA molecules. The FANTOM project, starting in early 2000, is one of the longest running collaborative projects in genomics that aims to functionally characterize mammalian genomes.79, 80 FANTOM1 and FANTOM2 generated more than 100 000 mouse full-length cDNAs that showed that the portion of the genome encoding proteins is small, whereas the majority of it is involved in producing non-coding RNAs. FANTOM3 uncovered the promoter landscape of the mammalian genome, allowing precise identification of TSS, and the existence of antisense transcription, by combining CAGE and full-length cDNA isolation.80 FANTOM4 adopted CAGE and 454 Life Sciences sequencing combined with Illumina microarrays to study a model of differentiation in human THP-1 (myeloid leukemia) cells, and to define the transcriptional regulatory network based on TSS activity that explained such a timely process.79 Within the FANTOM5 project, the consortium profiled nearly 2000 human and 1000 mouse samples, representative of the majority of cell types and tissues, using CAGE followed by next-generation single molecule sequencing (HeliScope).81, 82 A current limitation of FANTOM5, besides the coverage of species, lies in the approaches taken to explore RNAs. As the CAGE protocol is designed to capture only the 5′-end of capped long RNA molecules, the internal structure of long RNAs and small regulatory RNAs remains unexplored. To complement the CAGE profiles, CAGEscan, RNA-seq and small RNA sequencing data are being analyzed to be added to the FANTOM web resource, and existing databases and interfaces are being upgraded. The consortium is already developing the sixth FANTOM project which aims to uncover the function of long non-coding RNAs by high-throughput screening coupled with CAGE.
FANTOM projects revealed that the human genome is widely transcribed, producing thousands of lncRNAs. Most lncRNAs lack typical signatures of selective constraints. Given their diversity in biogenesis and their low expression and conservation levels, the functional relevance of most lncRNAs remains unclear. Future studies will enable understanding of lncRNA functions and to identify relevant lncRNA in diseases including prostate cancer.
Conclusion
The main priority in clinical prostate cancer research is identifying novel biomarkers to reliably distinguish between low-risk patients and high-risk patients who need definitive treatment. Strong evidence has been presented about the significance of lncRNAs in prostate cancer, and the challenge is now to determine lncRNAs functionally relevant with oncogenic changes. Although recent studies showed that measuring these transcripts in clinical samples could improve the prediction of prognosis, identification of more molecular mechanisms that lead to metastatic prostate cancer would enable us to determine treatment modalities at the time of diagnosis.
Disclosure Statement
Authors declare no conflicts of interest for this article.
Abbreviations
-
- ADT
-
- androgen deprivation therapy
-
- AR
-
- androgen receptor
-
- ARBS
-
- androgen receptor binding site
-
- ARE
-
- androgen responsive element
-
- AS
-
- antisense
-
- CAGE
-
- cap analysis of gene expression
-
- ChIP-seq
-
- chromatin immunoprecipitation sequencing
-
- CRPC
-
- castration-resistant prostate cancer
-
- DRE
-
- digital rectal examination
-
- EZH2
-
- enhancer of zeste homolog 2
-
- ISH
-
- in situ hybridization
-
- KLK3
-
- kallikrein-related peptidase 3
-
- lncRNAs
-
- long non-coding RNAs
-
- miRNAs
-
- microRNAs
-
- ncRNAs
-
- non-coding RNAs
-
- nt
-
- nucleotide
-
- PCA3
-
- prostate cancer antigen 3
-
- PSA
-
- prostate-specific antigen
-
- RNA-seq
-
- RNA sequencing
-
- TLR
-
- Toll-like receptor
-
- TTS
-
- transcriptional stat site




