CIC/ATXN1-rearranged tumors in the central nervous system are mainly represented by sarcomas: A comprehensive clinicopathological and epigenetic series
Arnault Tauziède-Espariat and Azadeh Ebrahimi share first authorship.
Abstract
CIC fusions have been described in two different central nervous system (CNS) tumor entities. On one hand, fusions of CIC or ATXN1 genes belonging to the same complex of transcriptional repressors, were reported in the CIC-rearranged, sarcoma (SARC-CIC). The diagnosis of this tumor type, which was recently added to the World Health Organization (WHO) Classification of CNS tumors, is difficult mainly because the data concerning its histopathology (as compared to its soft tissue counterpart), immunoprofile, and clinical as well as radiological characteristics are scarce in the literature. On the other hand, a recent study, based on DNA-methylation profiling, has identified a novel high-grade neuroepithelial tumor characterized by recurrent CIC fusions (HGNET-CIC). The aim of this multicentric study was to characterize a cohort of 15 primary CNS tumors harboring a CIC or ATXN1 fusion in terms of clinical, radiological, histopathological, immunophenotypical, and epigenetic characteristics. According to the integrated diagnoses, 14/15 tumors corresponded to SARC-CIC, and only one to HGNET-CIC. The tumors showed similar clinical (mainly pediatric), radiological (mostly supratentorial, cystic, and contrast enhancing), immunophenotypical (common expression of glioneuronal markers), and genetic (similar spectrum of fusions) profiles but their histopathological appearance was clearly distinct. Moreover, we found a novel fusion transcript (CIC::EWSR1) in a SARC-CIC. Most DNA methylation profiles using the Heidelberg Brain Tumor Classifier (v12.8) annotated the samples to the methylation class “SARC-CIC” (9/14 tumors with available data). By using uniform manifold approximation and projection analysis, four other samples were classified as SARC-CIC and another clustered within the methylation class of HGNET-CIC. Our findings confirm that CNS CIC-fused tumors do not represent a single molecular tumor entity. Further analyses are needed to characterize HGNET-CIC in more detail. These results may help to refine the essential diagnostic criteria for SARC-CIC and their terminology (with a suggested consensual name of sarcoma, CIC/ATXN1-complex rearranged).
1 INTRODUCTION
The latest World Health Organization's (WHO) Classification of Central Nervous System (CNS) Tumors, published in 2021, considerably modified the chapter on mesenchymal, non-meningothelial tumors [1, 2]. It included novel tumor types, such as the CIC-rearranged sarcoma (SARC-CIC) which was already known in the soft tissue [3, 4]. Based on their similar histopathology, the literature seems to argue in favor of the same tumor type occurring in two different anatomical locations. Thus, this diagnosis constitutes a novel challenge for neuropathologists who are unfamiliar with this rare and newly defined mesenchymal neoplasm. Within the CNS, the clinicopathological and radiological data for SARC-CIC remains scarce in the literature, limited to case reports or small series (n = 31) [5-18]. Moreover, CIC-fused CNS tumors seem to present particularities compared to their soft tissue counterparts in terms of phenotype (a glioneuronal differentiation/immunoprofile has been reported in CNS tumors) [8, 14, 15, 18] and genotype. Whereas a CIC::DUX4 fusion is encountered in 95% of SARC-CIC of the soft tissues, the molecular spectrum of CNS tumors seems to be broader with different fusion partners (NUTM1, LEUTX, DUX4) which may or may not include the gene CIC [5-18]. ATXN1 fusions are exceptional in soft tissues [18, 19]. Furthermore, to make things more complex, an epigenetically defined high-grade neuroepithelial tumor with CIC fusion (HGNET-CIC) has been recently identified, harboring a recurrent CIC::LEUTX fusion, [20]. Very few clinicopathological data are available for this recent entity. As a result, the essential diagnostic criteria for SARC-CIC defined by the current WHO classification ([1] evidence of a CIC gene fusion and [2] predominant round cell phenotype; and [3] mild nuclear pleomorphism; and [4] variable admixture of epithelioid and/or spindle cells; and [5] variably myxoid stroma; and [6] variable CD99 and frequent ETV4 and WT1 expression) are no longer up-to-date and need to be reevaluated [1]. The aims of our work were to extensively characterize a cohort of CNS tumors from the CIC-fused spectrum in terms of clinical data, imaging, histopathology and molecular biology, and to try to specify precise diagnostic criteria.
2 METHODS
2.1 Sample collection
The tumor samples and patients' retrospective clinical data were provided by the consultation archive database (1982–2023) from the neuropathology department at GHU Paris-Psychiatry and Neurosciences Sainte-Anne Hospital and by French expert centers within the RENOCLIP-LOC network (n = 11). Four additional patients were added from the Institute of Neuropathology, University of Bonn Medical Center in Germany. Case selection was based on the primary CNS tumors with a CIC rearrangement evidenced by Fluorescent in situ hybridization (FISH) analysis and/or with a CIC or ATXN1 fusion. The tissue analyses were performed in accordance with local ethics regulations. Four cases were previously reported in the literature (#11 in [20], #12 in [15], #14 in [11], and #15 in [6]).
2.2 Clinical and radiological data
The following clinical data were acquired for each patient when possible: sex, age at imaging diagnosis, age at pathological diagnosis, past medical history, clinical presentation, quality of resection, radiotherapy, chemotherapy, event-free survival (EFS) and overall survival (OS). When available, a central radiological review by experienced neuroradiologists (VDR, and NB) was performed. The following features were evaluated: location, size, diffusion signal and apparent coefficient diffusion (ADC), contrast enhancement, presence of cysts, necrosis, edema, and perfusion parameters.
2.3 Histopathological review and immunohistochemical analyses
A representative paraffin block was selected for each tumor. Hematoxylin-Phloxin-Saffron (HPS)-stained slides for all samples underwent central review by an experienced neuropathologist (ATE). Histopathological patterns were labeled as oligodendroglial-like, astrocytic-like, ependymoma-like (with pseudorosettes), small round cells, epithelioid, reticular, rhabdoid, spindle cells, and microcystic. Necrosis, microvascular proliferation, Rosenthal fibers, eosinophilic granular bodies, ganglion cells, neuropil islands, microcalcifications, siderophages (deposition of iron pigment in macrophages suggesting a past hemorrhage), perivascular lymphocytic infiltrates, fibrotic or myxoid changes were noted as present or absent. The tumoral pattern was assessed as circumscribed, diffuse, or both, based on the histopathology for instance the presence of entrapped neurons in the tumor and by using neurofilament staining. The mitotic index was monitored using 10 high-power fields (HPF; corresponding to 3.2 mm2) in a hot-spot area and was counted by two neuropathologists. Reticulin staining was also performed.
Unstained 3-μm-thick slides of formalin-fixed, paraffin-embedded tissues were obtained and submitted for immunostaining. The following primary antibodies were used: ETV4 (1:100, clone PEA3, Santa Cruz Biotechnology; Dallas, USA), NUT (1:100, clone C52B1, Sigma-Aldrich, Saint-Louis, USA), Glial Fibrillary Acidic Protein (GFAP) (1:200, clone 6F2, Dako, Glostrup, Denmark), OLIG2 (1:3000, clone C-17, Santa Cruz Biotechnology, Dallas, USA), vimentin (1:800, clone V9, Dako, Glostrup, Denmark), epithelial membrane antigen (1:200, clone GM008, Dako, Glostrup, Denmark), CKAE1AE3 (1:800, clone AE1AE3, Dako, Glostrup, Denmark), CD99 (1:10, clone 12E7, Dako, Glostrup, Denmark), S100 (1:200, clone 6F2, Dako, Glostrup, Denmark), SOX10 (1:200, clone IHC010, Diagomics, Blagnac, France), Neurofilament Protein (1:100, clone 2F 11, Dako, Glostrup, Denmark), NeuN (1:1000, clone A60, Sigma-Aldrich, Saint-Louis, USA), CD56 (pre-diluted, clone 123C3, Dako, Glostrup, Denmark), synaptophysin (1:150, clone DAK-SYNAP, Dako, Glostrup, Denmark), alpha-smooth muscle actin (1:6000, clone S100, Dako, Glostrup, Denmark), desmin (1:200, clone D33, Dako, Glostrup, Denmark), CD34 (1:40, clone Qbend10, Dako, Glostrup, Denmark), INI1 (BAF47) (1:50, clone 25/BAF 47, BD Biosciences, Franklin Lakes, USA), BRG1 (1:50, clone EPR3912, Abcam, Cambridge, United Kingdom), and Ki-67 (1:200, clone MIB-1, Dako, Glostrup, Denmark). External positive and negative controls were used for all antibodies. MIB-1 labeling index was estimated in a hot-spot area by two neuropathologists.
2.4 DNA methylation array processing and copy number profiling
Genomic DNA was extracted from fresh-frozen or formalin-fixed and paraffin-embedded (FFPE) tissue samples. DNA methylation profiling of all samples was performed using the Infinium MethylationEPIC (850 k) BeadChip (Illumina, San Diego, CA, USA) or the Infinium HumanMethylation450 (450 k) BeadChip array (Illumina) as previously described [21]. All computational analyses were performed in R version 3.3.1 (R Development Core Team, 2016; https://www.R-project.org). Copy-number variation analyses from 450 k and EPIC methylation array data were performed using the Conumee Bioconductor package version 1.12.0. Raw signal intensities were obtained from IDAT-files using the Minfi Bioconductor package version 1.21.4 [21]. Illumina EPIC samples and 450 k samples were merged into a combined data set by selecting the intersection of probes present on both arrays (combineArrays function, minfi). Each sample was individually normalized by performing a background correction (shifting of the 5% percentile of negative control probe intensities to 0) and a dye-bias correction (scaling of the mean of normalization control probe intensities to 10,000) for both color channels. Subsequently, a correction for the type of material tissue (FFPE/frozen) and array type (450 k/EPIC) was performed by fitting univariable, linear models to the log2-transformed intensity values (removeBatchEffect function, limma package version 3.30.11). The methylated and unmethylated signals were corrected individually. Beta-values were calculated from the retransformed intensities using an offset of 100 (as recommended by Illumina). All samples were checked for duplicates by pairwise correlation of the genotyping probes on the 450 k/850 k array. To perform unsupervised non-linear dimension reduction, the remaining probes after standard filtering [21] were used to calculate the 1-variance weighted Pearson correlation between samples. The following non-default parameters were applied: theta = 0, pca = F, max_iter = 20,000 perplexity = 10. Dimensionality reduction was then performed using the uniform manifold approximation and projection (UMAP) method (uwot R package) with the following non-default parameters: n_neighbors = 10, spread = 2, min-dist = 0.2.
3 RESULTS
3.1 Clinical and radiological characteristics
The patients' clinical and neuroradiological data are detailed in Table S1 and Figures 1 and 2 and Supplementary Figure 1. The median age of presentation in our cohort was 9.5 years (ranging from 0 to 40). Ten patients (67%) were children. The female-to-male ratio was 2.0 (10 females and 5 males). The tumors were mostly supratentorial (13/15) but two were cerebellar. Data from treatment and follow-up was available for 9/15 patients. A gross total resection was achieved in 6/9 patients with no residual disease as verified by the postoperative central neuroradiological review. Two patients underwent focal radiation therapy combined with chemotherapy, one patient received focal radiation therapy alone and two patients received only adjuvant chemotherapy. Four patients were free of disease at 1–60 months. Eight patients presented a local recurrence after the initial resection (mean EFS, 2 months; median EFS, 1 month), and six of them died rapidly after the recurrence (median OS, 17.5 months; mean OS, 18.5 months for the whole cohort).
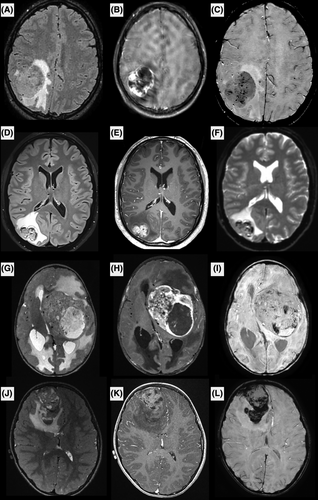
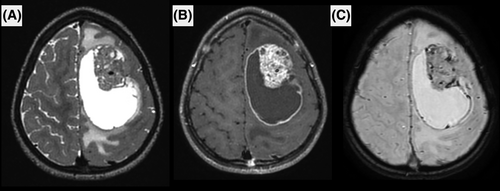
MRI was available for 9/15 patients, including pre- and post-contrast images, diffusion and perfusion imaging was available for seven. The images showed common features among the nine patients. All were large tumors (median largest diameter 52 mm, range [32–81]) with marked peritumoral edema (>1 cm) and intense contrast enhancement. Hemorrhage was seen in all cases, either as multiple microhemorrhages or gross tumor hemorrhage. Peripheral cysts with enhancing walls were seen in 8/9 patients, while microcysts within the tumor tissue were seen in 6/9 patients. Diffusion was restricted in 6/9, intermediate in 2/9 and uninterpretable for the remaining patient (median minimum relative ADC 0.98, IQR [0.82–1.14]). Perfusion was high in most cases using arterial spin labeling or dynamic susceptibility contrasting, except for one patient. All tumors were intra-axial, with 4/9 showing a large section of pachymeningeal contact, 3/9 showing limited pachymeningeal contact, and 1/9 cases showed only slight leptomeningeal contact. The last tumor had contact with the choroid plexus of the lateral ventricle without peripheral meningeal contact. None of the patients had brain metastases or spinal metastases when spinal MRI was available at diagnosis (3/9).
3.2 Histopathological and immunohistochemical characterization
The histopathological data of the patients are detailed in Table S1 and illustrated in Figures 3 and 4. All tumors were well-circumscribed from the brain or cerebellar parenchyma. Fourteen tumors presented similar compositions of dense sheets of tumor cells, with a variable amount of septa (often abundant) on silver impregnation, giving a vague lobular appearance. Using reticulin staining, there was no single cell ensheathment. A small round cells component was constantly observed and represented the predominant pattern in tumors admixed with spindle cells (7/14), a rhabdoid (5/14), or an epithelioid component (2/14). No ependymal (particularly pseudorosettes), astrocytic, oligodendroglial or glioneuronal features were present in those cases. No eosinophilic granular bodies, neuropil islands or perivascular inflammatory infiltrates were evident. Microcystic (5/14) and reticular (3/14) architecture were present. The stroma frequently presented myxoid changes (9/14), and one displayed a chondroid matrix. Hemorrhagic modifications were constantly present but hemosiderin depositions or siderophages were absent. The vascularization was well-developed with thin-walled branching vessels and focal microvascular proliferation in only two tumors.
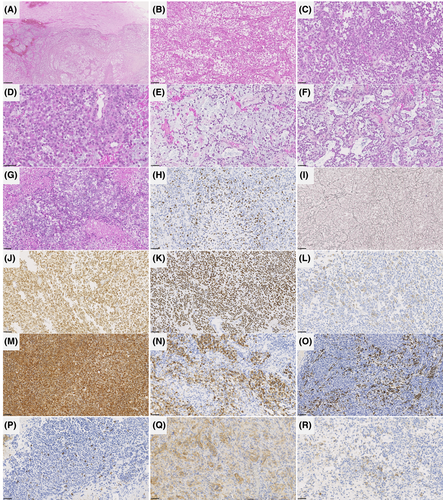
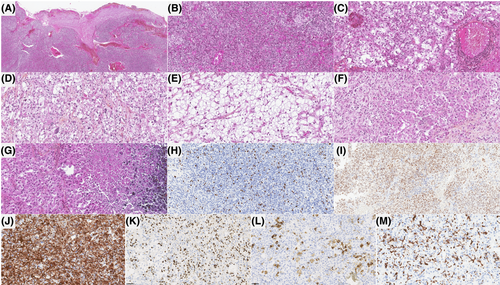
Necrosis was obvious in 10/14 cases and mitoses were frequent (11–31 per 1.6 mm2, with a mean of 22 mitoses). Ki-67 labeling activity was high, ranging from 15% to 80% (median: 30%). By immunohistochemistry, all tumors diffusely expressed vimentin. Epithelial (CKAE1/AE3 in one case, and EMA in four tumors), muscular (smooth muscle actin and desmin in two cases), neuronal (10/14), and glial (GFAP in 7/14 tumors, and OLIG2 in 5/14 cases) markers were variably expressed. INI1 and BRG1 immunoexpressions were constantly retained. A membranous staining for CD99 was present in 11/13 tested samples, mainly focal. ETV4 was constantly expressed whereas NUT immunopositivity was observed in five tumors.
One case (#14) was clearly distinct, having a polymorphous morphology composed of a predominant glioneuronal pattern with eosinophilic granular bodies, perivascular inflammatory infiltrates, CD34 extravascular cellular staining, GFAP, OLIG2 and synaptophysin expression in a large part of the tumor cells, admixed with clear cells, small round cells, and focal epithelioid cells. By immunohistochemistry, tumor cells also expressed vimentin, and ETV4. However, there was no immunoexpression for epithelial and myogenic markers. CD99 was focally expressed in this tumor. The silver impregnation showed a single cell ensheathment. Necrosis was obvious and mitoses were frequent (21 per 1.6 mm2). Ki-67 labeling activity was high (40%).
3.3 Genetic and epigenetic features, and integrated diagnoses
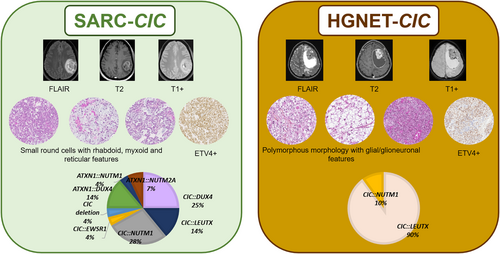
Alterations of the CIC gene were present in 12/15 cases, including: CIC::DUX4 (n = 5), CIC::NUTM1 (n = 4), CIC::LEUTX (n = 2), and CIC::EWSR1 (n = 1). The three remaining tumors presented an ATXN1::DUX4 (n = 2), and an ATXN1::NUTM1 (n = 1) fusion. First, DNA-methylation profiles were generated from the 14 tumor samples that had sufficient DNA quality. Using DNA methylation-based classification, nine of the tumors were classified as SARC-CIC (calibrated scores for DNA MC ≥0.9), and seven of them were shown to harbor a CIC fusion. Four other samples were classified as SARC-CIC but with a low calibrated score (<0.9), and the last case was not classified. Next, we performed an UMAP analysis, including the recently described MC of HGNET-CIC [20], of the whole cohort to better classify the tumors with low calibrated scores (<0.9). Thirteen tumors clustered with SARC-CIC reference tumors. Because they presented similar histopathological characteristics, the integrated diagnosis of each was SARC-CIC. The remaining case (#14) grouped within the class of HGNET-CIC reference tumors (Supplementary Figure 2). As it was morphologically different from the other tumors in our series, we considered an integrated diagnosis of HGNET-CIC.
No recurrent copy number variations were observed in the two subgroups (Supplementary Figure 3).
4 DISCUSSION
In the CNS, two different entities are now known to harbor recurrent CIC/ATXN1 fusions: the newly WHO CNS 2021 defined SARC-CIC and the recently described HGNET-CIC [5, 20] (Figure 5). In the current work, our comprehensive detailed study includes clinical, radiological, histopathological and genetic data from a series of CNS tumors with CIC spectrum fusions, mainly represented by SARC-CIC. They presented frequently as a small round cell morphology admixed with variable epithelioid, rhabdoid or spindle cells. As previously reported, SARC-CIC constantly expressed ETV4 [22] and may express the protein NUT for those harboring a NUTM1 fusion [5]. Surprisingly, these sarcomas may express markers from different lineages (epithelial, myogenic, glial and neuronal), which explains why some of them were initially diagnosed in the literature as high-grade gliomas or embryonal tumors [5, 7, 8, 15]. The expression of neuronal and epithelial markers has also been reported in SARC-CIC of soft tissues [23-27], but to our knowledge, no data are available about glial expression in extra-CNS tumors. As was observed in the current series, SARC-CIC mainly affect children and young adults (median age: 8 years), with 70% of patients being found in the pediatric age group [5-18]. There is no gender predisposition (sex ratio male/female of 1.1) in the literature [5-18]. Most SARC-CIC of the CNS occur in supratentorial sites (85% of reported tumors) [37–46] whereas spinal and posterior fossa (the two first examples are in the current series) presentation accounts for 10% and 5% of reported cases, respectively [5-18]. For the first time, we showed that SARC-CIC may present recurrent imaging features. These are characterized by large intra-axial tumors with frequent meningeal contact, marked peritumoral edema, intense contrast enhancement, hemorrhagic foci, diffusion restriction, and high perfusion. Because of the age of onset, morphology and pluriphenotypic immunoprofile, the main differential diagnosis for SARC-CIC is atypical teratoid and rhabdoid tumors (AT/RT), which are easily ruled out by immunohistochemistry (showing a retained expression of INI1 and BRG1). Contrary to their soft tissue counterparts, the molecular spectrum of CNS tumors (including our data) seems to be wider with different fusion partners for CIC and more frequent ATXN1 fusions [5-8, 11-15, 17, 18]. In this study, we reported for the first time a CIC::EWSR1 fusion. Our study confirmed that CIC-fused and ATXN1-fused sarcomas presented histopathological and epigenetic similarities, clustering together within the same MC [12, 13, 15, 18]. CIC or ATXN1 genes belong to the same complex of transcriptional repressors. ATXN1 is a Chromatin-binding factor that represses Notch signaling in the absence of Notch intracellular domain by acting as a C-repeat/DRE binding factor 1 (CBF1) corepressor and in concert with CIC involved in brain development [28]. The current series confirmed that DNA-methylation proven SARC-CIC may present the CIC::LEUTX fusion, which has been recurrently observed in HGNET-CIC [20]. Because HGNET-CIC shares clinical (young age), radiological (presented here in only one tumor) and molecular features (mainly CIC::LEUTX fusions, but also a CIC::NUTM1 fusion in one reported tumor) with SARC-CIC, the question of a same tumor type presenting a wide spectrum of morphology from pure mesenchymal to glioneuronal or subtype may be considered. However, to our knowledge, no SARC-CIC with glial or glioneuronal morphological features has been identified in the literature or in this cohort. The expression of glial and neuronal markers may only represent the pluriphenotypic pattern of these tumors, which is also the case with other CNS tumors, such as AT/RT. According to the original description [20], HGNET-CIC present a different morphology with a predominant glial and/or glioneuronal component (oligodendrocyte-like features, glial processes, pseudorosettes, eosinophilic granular bodies) and a distinct DNA-methylation profile [20]. With regards to the literature and the current case, it seems that HGNET-CIC are characterized by a pleomorphism of morphological patterns, similar to what one might observe in pleomorphic xanthoastrocytomas (PXA) or supratentorial ependymomas, ZFTA-fused for example [29]. Like SARC-CIC, we found that the case of HGNET-CIC expressed ETV4 and CD99. In these conditions, ETV4 would be an interesting marker for screening the CIC fusions in the spectrum of high-grade glial/glioneuronal tumors, IDH, H3- and BRAF-wildtype. In the previous study, Sievers et al. suggested vimentin as a potential differential marker to distinguish sarcomas from HGNET-CIC [20]. However, our experience has shown vimentin to be diffusely expressed by both SARC-CIC and HGNET-CIC. Consequently, the diagnosis of HGNET-CIC remains to be a challenge, and the current epigenetic classifier of brain tumors does not include this MC. This diagnosis may integrate histopathology (glioneuronal tumor) and the presence of a CIC fusion. Contrary to SARC-CIC, it seems that HGNET-CIC are only supratentorial [20]. Like their soft tissue counterpart, most SARC-CIC follow an aggressive course with frequent recurrences (61% of patients), most commonly local, and death (45% of patients) [5-9, 11-16, 18]. In terms of prognosis, further series of HGNET-CIC are needed to determine their behavior (data are available for only six patients, mean progression-free survival: 13.5 months and only one patient died 15 months after the initial diagnosis) [20].
In the current WHO Classification, the DNA-methylation profile constitutes a desirable criterion for the diagnosis of SARC-CIC in the CNS. For laboratories that do not have access to the methylation-based classification, FISH CIC can be used as an important diagnostic tool for this tumor type.
To conclude, SARC-CIC and HGNET-CIC may share similar fusions, clinical and radiological findings, and the same immunoprofile. Further studies are needed for a detailed histopathological characterization of HGNET-CIC to confirm if they represent a novel pleomorphic glial/glioneuronal tumor entity rather than a subtype or variant of SARC-CIC. This work and the recent literature have evidenced that the current terminology for CIC-rearranged sarcoma is too restrictive and the essential criteria should be improved. Because SARC-CIC may present non-CIC fusions, we suggest CNS sarcoma, CIC/ATXN1 complex-fused, as a closer consensual nosological term for this group of tumors. In addition, we suggest the following diagnostic criteria: a mesenchymal high-grade primary CNS tumor having a predominant pattern of small round cell morphology; AND INI1 and BRG1 retained expressions; AND ETV4 expression; AND the presence of a CIC or ATXN1 fusion.
AUTHOR CONTRIBUTIONS
A.T.E., A.S., E.U.C., F.B., K.M., A.M.E., M.S., C.A.M., B.L., A.E., W.G., T.B., A.K., O.A., J.B., D.C., C.G., and P.V. conducted the histopathological examinations. P.S., D.G., Y.N., A.M., and G.P. conducted the molecular studies. A.T.E., L.H., and P.V. drafted the manuscript; V.D.R., A.G., and N.B. reviewed all imaging data. V.L., G.I., T.B., and K.B. recruited patients, provided samples, and clinical informations. All authors reviewed the manuscript.
ACKNOWLEDGMENTS
We would like to thank the laboratory technicians at GHU Paris Neurosciences, Hospital Sainte-Anne, for their assistance.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data supporting this study are available from the corresponding author upon reasonable request.




