Time-resolved single virus tracking and spectral imaging to understand HIV-1 entry and fusion
Abstract
Single Virus Tracking (SVT) is a key technique to understand how individual viral particles evolve during the infection cycle. In the case of the human immunodeficiency virus (HIV-1), this technology, which can be employed using a simple and affordable wide-field microscope, has proven to be very useful in the first steps of infection, such as the kinetics of the fusion reaction or the point of fusion within live cells. Here, we describe how SVT in combination with other spectral imaging approaches is a powerful technique to illuminate crucial mechanistic aspects of the HIV-1 fusion reaction. We also stress the role of our laboratory in elucidating a few mechanistic aspects of retroviral fusion employing SVT such as: (i) the role of dynamin, (ii) how metabolism modulates membrane composition and cholesterol and its impact in fusion, (iii) the importance of envelope glycoprotein (Env) intra- and inter-molecular dynamics for neutralization, or (iv) the time-resolved fusion stoichiometry in three characteristic steps for the HIV-1 prefusion step. These observations constitute a good testimony of the complexity of retroviral fusion and show the strength of SVT when applied to live cells and combined with quantitative spectral approaches. Finally, we propose several crucial remaining questions around HIV-1 fusion and how the combined use of these technologies, always in live cells, will be able to shed light into the intricacies of arguably the most important step of the HIV-1 infection cycle.
INTRODUCTION
The Human Immunodeficiency Virus-1 (HIV-1) infects target cells by priming its envelope glycoprotein (Env) with the CD4 receptor and co-receptors (either CCR5 or CXCR4). After engagement between Env and CD4 and co-receptor interactions, there is a series of conformational changes that facilitate virus cell fusion and the formation of the gp41 post-fusion stable six helix bundle (Roben et al., 1994). Even if this process has been studied thoroughly during the last 4 decades, there are several questions unanswered (Jakobsdottir et al., 2017). For instance, which are the molecular mechanisms that facilitate productive fusion? How intra- and inter-molecular dynamics of Env are engaged with CD4 and co-receptors? How are these processes coordinated with other host cell factors including lipids?
To solve the mystery of this highly dynamic process, a lot of effort has been put into elucidating Env structure alone (Lee et al., 2016) and when engaged to CD4 (Huang et al., 2016) or other ligands such as broadly neutralizing antibodies (bNAbs) (Wu et al., 2010). In these structures, Env adopts open, partially open, and prefusion conformations rather than a single stable conformation. This has been corroborated by the Mothes group using single molecule Forster Resonance Energy Transfer (smFRET) experiments in HIV-1 Env particles (Lu et al., 2019; Munro et al., 2014) and by our own group in both HIV-1×4 tropic viruses in vitro and when engaged in T cells (Carlon-Andres et al., 2021). The information that comes from time-resolved single virus tracking and its combination with spectroscopy and other correlative approaches in live cells has been crucial for increasing our knowledge of virus entry. For instance, human immunodeficiency virus (HIV-1) fusion has been shown to depend on the lipid composition of both the virus and the host (Coomer et al., 2020). The caveats of these and other technologies, such as super-resolution, based on light microscopy are mainly the labeling strategies and to a lesser extent the spatial resolution. Whichever method is employed to label different regions of the target protein in both the virus and the host, loss of functionality is the key parameter that needs to be carefully monitored. Here we review how SVT alone or when combined with other light microscopy approaches has unveiled biological mechanisms that drive the HIV-1 fusion reaction. This integrative approach, which considers both the virus and the host has great potential to increase our knowledge relative to the many factors that control HIV-1 productive fusion.
IMAGING VIRUS PRIMING AND FUSION
Real-time single virus tracking on live cells: Unraveling the role of Dynamin in HIV-1 fusion
Real-time Single Virus Tracking (SVT) is a powerful tool to understand virus entry (Maldonado et al., 2020; Padilla-Parra et al., 2014). By labeling different HIV-1 viral compartments with different fluorophores, one can track in time and space single viruses as they enter the cell. The use of wide-field fluorescence microscopy equipped with modern cameras (such as EM-CCD or sCMOS) provides good time resolution to be able to follow double-labeled particles (Figure 1). Other approaches such as total internal reflection microscopy (TIRFM) are also adequate for HIV-1 particles engaging with CD4 and coreceptors and if used in Hi-Lo mode could also help to visualize endosomal entry (Jones et al., 2017; Padilla-Parra et al., 2014). The development of new photon-counting hybrid detectors (digital detectors) for confocal microscopy also permitted the use of confocal microscopy for real-time SVT imaging of live cells (Russell et al., 2017).
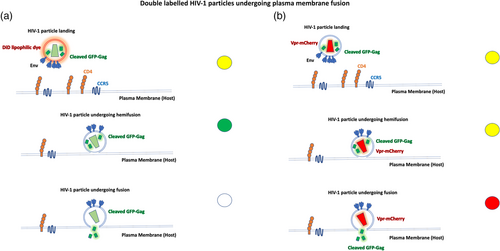
The Melikyan, Hope laboratories pioneered the use of double-labeled HIV-1 viruses to quantify single virus fusion in live cells (Markosyan et al., 2005; McDonald et al., 2002). Until then, suitable assays to follow both lipid and contents mixing, step by step did not exist. Briefly, the dissection of individual fusion events into quantifiable discrete steps was achieved by utilizing a lipophilic dye in the envelope (such as the far-red dye DiD) and Gag-GFP, which is naturally cleaved to NC-GFP fragments in mature particles. Upon fusion opening, the NC-GFP fragment is released to the cytosol while the capsid is retained. The lipid marker DiD is transferred from HIV-1 particles to cells upon hemifusion in the plasma or endosomal membrane. (Figure 1a). This ingenious labeling approach has been employed ever since in many scenarios(de la Vega et al., 2011; Miyauchi et al., 2009; Padilla-Parra et al., 2013; Russell et al., 2017) and has inspired other labeling approaches that follow the same logic employing other fluorescent markers, such as quantum dots, or click chemistry (Li, Yin et al., 2018). Other labeling approach was employed labeling Vpr with mCherry (Vpr-mCherry) and Gag with GFP (Gag-GFP) (Figure 1b). In this instance the double labeled particles change from yellow (Gag-GFP + Vpr-mCherry) to red upon fusion (Padilla-Parra et al., 2013). Another approach consisted in introducing other genetically encodable fluorescent proteins (FP) in the core of HIV-1 particles (Padilla-Parra et al., 2013). HIV-1 pseudoviruses were co-labeled with Gag-iCherry and YFP-Vpr. Two different FPs with different pKa (mCherry has a low pKa of 4.5 while eYFP has a pKa of 6.9) permitted to detect both HIV-1 fusion and intraviral pH. Inspired by the FRET-based pH sensor developed by the Wouters group termed pHlameleons (Esposito et al., 2008), we built a modified version of the original pH nanosensors (ECFP-linker-eYFP) into ICAM-1 (ICAM-1-mTFP1-eYFP) which is known to be incorporated in the envelope of retroviruses (both HIV-1 and avian sarcoma and leukosis virus (ASLV)) (Padilla-Parra et al., 2012). The strategy here, was to quantitatively measure the pH of single ASLV particles while getting endocytosed also utilizing a third marker for the core (Gag-mKate2) to quantify the endosomal pH from acidification to full fusion in a retrovirus known to possess a pH-dependent fusogen. Cabot et al. (2022) replicated this approach using another pHlameleon (ICAM-1-mTFP1-mCitrine). Interestingly, other pH biosensors incorporated in retroviral particles have also been explored with success to determine the delay between pH endosomal acidification and full retroviral fusion (Padilla-Parra et al., 2014).
The use of SVT together with other functional assays for HIV-1 fusion and pharmacological intervention led Miyauchi et al. (2009) to suggest that Dynamin played an important role in HIV-1 fusion and that HIV-1 enters the cell via endocytosis. Also, SVT and different functional assays (time-of-addition beta-lactamase assay (Carlon-Andres & Padilla-Parra, 2020; Cavrois et al., 2002; Jones & Padilla-Parra 2016) showed the importance of dynamin and endocytosis during HIV-1 fusion. The exact role of dynamin-2 during HIV-1 entry was determined employing several imaging approaches and introducing for the first time SVT and Number and Brightness (NandB) (Digman et al., 2008) combined with Total Internal Reflection Microscopy (TIRF). Here, it was shown that the mechanical process of fusion pore formation and enlargement needs the orchestrated role of several molecular partners (Jones et al., 2017), being dynamin 2 one of them. Using both TZM-bl reporter cells and primary CD4+ T cells, it was hypothesized that dynamin-2 plays a multifaceted role during HIV-1 fusion reaction. First, it senses curvature and helps to drive hemifusion preventing fission from happening. Second, dynamin-2, with a low oligomeric state (probably a tetramer) is able to stabilize the fusion pore right after hemifusion. Interestingly, Aggarwal et al. were able to corroborate these observations by performing SVT and fusion functional assays also in other T cell lines (Aggarwal et al., 2017).
IMAGING THE PHYSIOLOGICAL STATE OF TARGET CELLS DURING VIRUS ENTRY USING FRET-BASED BIOSENSORS COMBINED WITH SVT
SVT combined with genetically encodable biosensors; the role of cholesterol regulation and metabolism during HIV-1 entry
The combination of SVT with fast fluorescence lifetime imaging microscopy (FLIM) (Padilla-Parra et al., 2015) allowed to unveil another mechanistic aspect that modulates HIV-1 entry and fusion: metabolism and cholesterol concentration in the plasma membrane (Coomer et al., 2020). In the past, FLIM suffered from long acquisition times and complex mathematical approaches for data analysis. Recent developments in (i) digital detectors allowed reducing the deadtime of these HyD detectors together with (ii) fastest electronics and (iii) non-fitting approaches (Padilla-Parra et al., 2011). These technological advancements have rendered FLIM to a broader audience of biologists (Leray et al., 2013). This is very important in the field of biosensors, as FLIM permits accurate quantification of Forster resonance energy transfer (FRET) regardless the optical path or the level of expression in different subpopulations of cells. Taking advantage of these new features in technology and analysis we combined SVT with genetically-encodable biosensors such as Perceval (Berg et al., 2009) which measures the ratio of ATP/ADP; Laconic that measures the relative concentration of lactate (San Martín et al., 2013) or MSS that measures membrane tension (Li, Yu et al., 2018). SVT, allowed to monitor how cell metabolism modulates HIV-1 fusion in both TZM-bl reporter cells and primary CD4+ T cells (Coomer et al., 2020). Using lifetime imaging (FLIM), time-resolved data from single HIV-1 particles could be gathered with information on the microenvironment sensed by individual viruses. Here, it was shown that inhibition of glycolysis sequesters cholesterol from the plasma membrane in both TZM-bl cells but also primary T cells. The metabolic landscape of individual cells turned out to determine the propensity for productive HIV-1 fusion (Coomer et al., 2020). The role of cholesterol in HIV-1 infection has long been investigated (Serquiña et al., 2017). It has been shown that the conditions for CD4+ T cells depends on the metabolic program (Valle-Casuso et al., 2019). Indeed, the metabolic profile was shown to be a crucial factor on infection of CD4+ T cells; however, in this work, it was not precisely determined how metabolism could affect HIV-1 entry and fusion. Other work showed that high glycolytic activity may increase susceptibility to different cell types (MT4, primary T cells, or TZM-bl) and support HIV-1 entry and fusion due to membrane tension regulation (Coomer et al., 2020). In this context, cholesterol sequestration followed by glycolytic inhibition was specific and perhaps regulated through acetyl-CoA precursors, as other receptors such as CD4 and CCR5 or CXCR4 expression levels remained unperturbed during 2 deoxyglucose (2DG) exposure in both TZM-bl and MT4 T cells (Coomer et al., 2020). Thanks to these combined measurements employing genetically encodable biosensors and SVT, it was determined how the local plasma membrane tension drops right at the moment of full fusion. This observation was further supported by the use of FLIP-TR membrane order probe originally developed by the Roux lab (Colom et al., 2018). TZM-bl reporter cells were exposed to HIV-1 viruses decorated with JR-FL Env and different concentrations of 2-DG. Different phenotypes for membrane order were found depending on cholesterol sequestration from the plasma membrane, which in turn correlated with HIV-1 fusion and infection. Importantly, individual HIV-1 viruses (with both X4 and R5 tropic Env) would not be able to progress from hemifusion in cells were glycolysis was disrupted. Moreover, based on these observations, Coomer et al. hypothesized that there must be a hydrophobic mismatch between ordered and disordered lipid domains that renders HIV-1 fusion possible.
MULTIPLEXING ADVANCED FLUORESCENCE MICROSCOPY TECHNIQUES WITH SVT
FRET-FLIM and single virus tracking in live cells: Disentangling inter- and intra-molecular Env dynamics when primed to T cells
The combination of lifetime imaging employed to detect Forster resonance energy transfer (FRET-FLIM) and SVT allowed to probe intra and inter HIV-1 Envelope glycoprotein (Env) molecular dynamics of single HIV-1 viruses alone and when primed to live T cells (Carlon-Andres et al., 2021). As explained above, FRET-FLIM allows to probe protein-protein interactions with high accuracy and is extremely useful when applied in live cells (Carlon-Andres & Padilla-Parra, 2020). These dynamics were also tested in the presence of inhibitory concentrations of broadly neutralizing antibodies (bNAbs) targeting different Env epitopes. Regardless of the Env target of these bNabs (e.g., the apex, the CD4 binding region, or the membrane proximal region (MPER)) all antibodies turned out to disrupt Env intermolecular dynamics while simultaneously stabilizing an open or closed Env conformation as seen by others (Ma et al., 2018). Env intermolecular dynamics was first seen employing super-resolution approaches by others (Chojnacki et al., 2012). Chojnacki et al. observed that in mature HIV-1 particles Env was organized in groups or “clusters.” Even if stimulated light emission depletion (STED) nanoscopy could reach a spatial resolution of around 30–50 nm also seen by in HIV-1 viruses (Carlon-Andres et al., 2021) one could not define the dynamic nature nor the function of these so-called cluster of Envs. The combination of FRET-FLIM and SVT permitted to highlight a potential previously unappreciated Env functionality only in matured HIV-1 particles (Figure 2a). The use of FRET-FLIM employing a 2D kernel plot or a phasor plot allowed to define different FRET profiles for different conformational Env states (both inter- and intra-molecular) (Figure 2a). The 2D kernel plot was obtained plotting all pixel values coming from two different images: The lifetime image (in ns) and the apparent FRET efficiency image. In the case of the Phasor Plot (Digman et al., 2008; Padilla-Parra et al., 2011), a graphical representation of all the lifetime imaging microscopy (FLIM) data in a vector space, also gave rise to different regions with a particular Env conformation. Based on three dimensional models of Env structures we allocated these FRET regimes to open, closed and inter Env interactions. These analyses were performed in both, single virus particles alone and when exposed in MT4 T cells (Figure 2a).
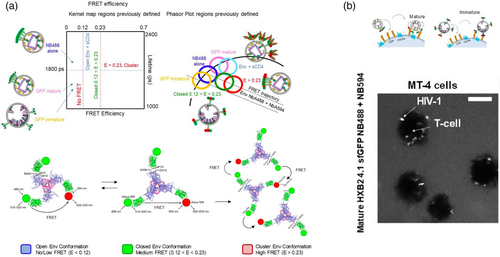
By producing immature particles utilizing protease inhibitors (Padilla-Parra et al., 2012), it was shown that intermolecular dynamics were not seen in FRET-FLIM nor in STED; corroborating previous studies where it was clearly established that Env tends to form groups only in mature particles (Chojnacki et al., 2012). Env grouping might be related to immune evasion and could be a necessary means for the virus modulate HIV-1 priming and fusion. Moreover, it is plausible that within these groups of Env the intramolecular dynamics might be affected offering an extra layer of complexity to rational vaccine design based on isolated SOSIP structures that turned out to produce partially open or fully open Env structures only (Lu et al., 2019). Importantly, these single molecule FRET-FLIM experiments were also performed when real HIV-1 particles were engaged in CD4+ T cells in the presence and absence of broadly neutralizing antibodies (Carlon-Andres et al., 2021) (Figure 2b). Finally, it is worth mentioning that within the 30–50 nm resolution realized by STED one could see clear differences only between mature and immature particles. FRET-based approaches, however, could also establish differences between mature viruses engaged with living T cells exposed or not to a number of broadly neutralizing antibodies (bNabs) during the first steps of the fusion reaction. It is possible that the higher FRET spatial resolution (10 nm) reflects intermolecular Env changes that would be unappreciated when employing STED microscopy.
Number and brightness and single virus tracking in live cells: Probing the time-resolved stoichiometry of the HIV-1 fusion reaction in live cells
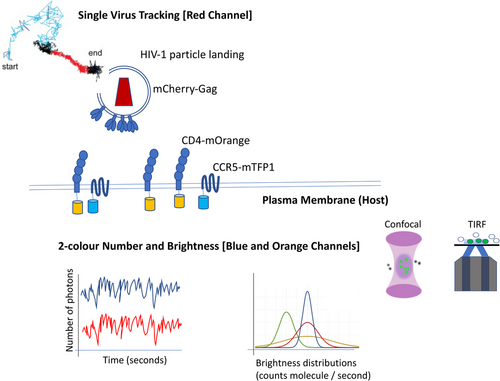
Even if inter and intramolecular Env dynamics is crucial for understanding neutralization mechanisms, one must look at the interplay between the host and the virus. The use of two-color Number and Brightness (Bcc) (Digman et al., 2009) in combination with SVT permitted following the stoichiometry of the pre-fusion reaction for single HIV-1 particles when engaged in live cells (Iliopoulou et al., 2018). Briefly, number and brightness (NandB) (Digman et al., 2008) is a technique that analyses the first and the second moments (average and variance) of the photon counting histogram (PCH) (Jiang et al., 2020) recovered from the fluorescence fluctuation trace that single molecules leave out when diffusing through a confocal volume (Nolan et al., 2018). This technique allows quantitation of the average oligomeric state of a particular fluorophore, and it can be employed in a pixel-by-pixel mode. Digman et al. also employed the two-color cross-correlation NandB (ccNandB or Bcc) to determine the stoichiometry of a particular interaction in live cells (Digman et al., 2009). The combination of NandB using two different channels and SVT with a single labelled virus allowed extracting the individual time traces for CD4-mOrange on one side and CXCR4-mTFP1 or CCR5-mTFP1 on the other to recover the stoichiometries of receptor and coreceptor interactions around single HIV-1 particles labeled with Gag-mCherry.
The combination of time-resolved two-color Bcc with SVT allowed to quantify the intermolecular dynamics of CD4 and co-receptors in permissive living cells during the HIV-1 pre-fusion reaction for both X4-tropic HIV-1 Env HXB2 strain and an R5-tropic HIV-1 primary isolate (JRFL) (Figure 3). Interestingly, immature particles were uncapable to progress in the pre-fusion reaction as no significant increase in CD4 or CXCR4 oligomeric states was observed. These results were also seen with R5 tropic HIV-1 particles. When exposing other ligands to live cells known to inhibit HIV-1 fusion such as b12, CD4 antibodies or co-receptor ligands, no positive cross-correlation between the CD4 and coreceptors was detected around mature HIV-1 particles. The stoichiometries obtained for both R5 and X4 tropic viruses differed: for X4 tropic viruses, an asymmetric complex was found when Env interacted with a single CD4 molecule (interestingly this complex was seen in both X4 and R5 tropic viruses). Right after CXCR4 was engaged with Env as a dimer (second step) and next recruitment of a second Env and six CD4 molecules took place (third step). This prefusion three-step mechanism occurred prior to six helix bundle formation and fusion. For R5 tropic viruses, it was also demonstrated a three-step prefusion mechanism where a single Env would interact with a single CD4 molecule (first step and formation of asymmetric complex). Concomitant recruitment of CCR5 dimer and two CD4 (second step) molecules would stabilize a single Env (1) and CD4 (3) CCR5 (2) complex (third step) prior to six helix bundle formation and fusion pore formation. These results based on the combination of fluorescence fluctuation spectroscopy (FFS) and SVT were also supported by multi-color single molecule localization microscopy. It is worth noting that the asymmetric intermediate (step 1) seen for both X4 and R5 tropic HIV-1 viruses, was also suggested by Kwon et al. (2015) employing crystallographic approaches and single molecule FRET.
What's next from the technological perspective?
The next step to apply SVT combined with spectral imaging would be to visualize inter- and intra-molecular Env dynamics from single HIV-1 particles interacting with endogenously labeled receptors and co-receptors in live T cells. This implies two degrees of complexity: first to produce a stable T cell line with endogenous labeled CD4 and CCR5 and/or CXCR4; second, to combine the two approaches described above FRET-FLIM and FFS with SVT. The third technological challenge would be to be able to recover at least 5–8 colors simultaneously to recover information on single HIV-1 particles on maturation, Env dynamics, and priming with T cell CD4 receptors and co-receptors.
CONCLUSIONS AND OVERLOOK
HIV-1 fusion is a time-resolved reaction with multifaceted and complex steps that are often-oversimplified. When dissecting this problem, one needs to consider single HIV-1 viral particles and the interplay with their host. Reductionistic and oversimplified in vitro approaches are necessary to a certain extent but should not constitute the crux of a rigid paradigm which often tends to ignore mechanistic and dynamic aspects during the fusion reaction. The structure of Env has received major attention over the last few decades and is one of the keys to understand HIV-1 fusion. Its highly dynamic nature and the many important factors discussed here suggests to change the paradigm and focus on a multifaceted approach in which to build the HIV-1 theory of membrane fusion. There are still many questions to be resolved in this limited model, but understanding how Env dynamics alter receptor stoichiometry, the role of lipids and how their regulation affect membrane structure and rigidity or how other host factors like actin regulation or dynamin-2, is the key to fully describe arguably the most important infection step of HIV-1.
AUTHOR CONTRIBUTION
Sergi Padilla-Parra conceptualized the review, and wrote the article and produced the figures.
ACKNOWLEDGMENT
We thank the members of the Padilla-Parra lab for constructive comments. This work has been supported by the European Research Council (ERC-2019-CoG-863869 FUSION to S.P.-P.).
CONFLICT OF INTEREST
The funders played no role in study design and analysis, the decision to publish, or preparation of the manuscript. We declare that we have no competing interests.
Open Research
DATA AVAILABILITY STATEMENT
All data are freely available and/or are included in the review.