Hepatitis B virus movement through the hepatocyte: An update
Abstract
Viruses are obligate intracellular pathogens that utilize cellular machinery for many aspects of their propagation and effective egress of virus particles from host cells is one important determinant of virus infectivity. Hijacking host cell processes applies in particular to the hepatitis B virus (HBV), as its DNA genome with about 3 kb in size is one of the smallest viral genomes known. HBV is a leading cause of liver disease and still displays one of the most successful pathogens in human populations worldwide. The extremely successful spread of this virus is explained by its efficient transmission strategies and its versatile particle types, including virions, empty envelopes, naked capsids, and others. HBV exploits distinct host trafficking machineries to assemble and release its particle types including nucleocytoplasmic shuttling transport, secretory, and exocytic pathways, the Endosomal Sorting Complexes Required for Transport pathway, and the autophagy pathway. Understanding how HBV uses and subverts host membrane trafficking systems offers the chance of obtaining new mechanistic insights into the regulation and function of this essential cellular processes. It can also help to identify potential targets for antiviral interventions. Here, I will provide an overview of HBV maturation, assembly, and budding, with a focus on recent advances, and will point out areas where questions remain that can benefit from future studies. Unless otherwise indicated, almost all presented knowledge was gained from cell culture-based, HBV in vitro-replication and in vitro-infection systems.
Abbreviations
-
- AGL
-
- antigenic loop
-
- AP
-
- adaptor protein
-
- ApoE
-
- apolipoprotein E
-
- ATF6
-
- activating transcription factor 6
-
- Atg
-
- autophagy proteins
-
- ARD
-
- arginine-rich domain
-
- CAD
-
- cytosolic anchorage determinant
-
- CCC
-
- covalently-closed-circular
-
- CDK2
-
- cyclin-dependent kinase 2
-
- CHB
-
- chronic hepatitis B
-
- COPII
-
- coat protein complex II
-
- CRM1
-
- chromosome region maintenance 1
-
- CYL
-
- cytosolic loop
-
- DN
-
- dominant-negative
-
- EndoH
-
- endoglycosidase H
-
- ER
-
- endoplasmic reticulum
-
- ERAD
-
- endoplasmic reticulum associated degradation pathway
-
- ERGIC
-
- ER-Golgi intermediate compartment
-
- ERGIC-53
-
- ER-Golgi intermediate compartment 53 protein
-
- ESCRT
-
- endosomal sorting complexes required for transport
-
- fSVP
-
- filamentous subviral particles
-
- HBcAg
-
- hepatitis B core antigen
-
- HBeAg
-
- hepatitis B e antigen
-
- HBsAg
-
- hepatitis B surface antigen
-
- HBV
-
- hepatitis B virus (HBV)
-
- HIV-1
-
- human immunodeficiency virus type 1
-
- Hrs
-
- hepatocyte growth factor-regulated tyrosine kinase substrate
-
- IFN-α
-
- interferon alpha
-
- Imp-α
-
- importin-α
-
- Imp-ß
-
- importin-ß
-
- IRE1α
-
- inositol-requiring protein-1α
-
- LMAN-1
-
- lectin mannose-binding 1 protein
-
- MAP1S
-
- microtubule-associated protein 1S
-
- MD
-
- matrix domain
-
- MLV
-
- murine leukemia virus
-
- MT
-
- microtubules
-
- MVB
-
- multivesicular body
-
- NAPs
-
- nucleic acid polymers
-
- NC
-
- nucleocapsid
-
- NES
-
- nuclear export signals
-
- NLS
-
- nuclear localization signals
-
- NPC
-
- nuclear pore complex
-
- NTCP
-
- sodium-taurocholate cotransporting protein
-
- Nup153
-
- nucleoporin 153
-
- NXF1
-
- nuclear export factor-1
-
- OST
-
- oligosaccharyltransferase
-
- ORF
-
- open reading frame
-
- PDI
-
- protein disulfide-isomerase (PDI)
-
- PG
-
- pregenomic
-
- Pin1
-
- peptidyl-prolyl cis-trans isomerase NIMA-interacting
-
- PLK1
-
- polo-like kinase 1
-
- PRMT5
-
- protein arginine methyltransferase 5
-
- RC
-
- relaxed circular
-
- RT
-
- reverse transcriptase
-
- sAPs
-
- secretory autophagosomes
-
- SERINC5
-
- serine incorporator 5
-
- SRPK1
-
- serine/arginine-rich protein kinase 1
-
- SRPK2
-
- serine/arginine-rich protein kinase 2
-
- sSVP
-
- spherical subviral particles
-
- SNAP-29
-
- synaptosomal-associated protein 29
-
- SNAREs
-
- soluble N-ethylmaleimide-sensitive attachment receptors
-
- SRP
-
- signal recognition particle
-
- STAM1/2
-
- signal transducing adaptor molecule1/2
-
- STOPS
-
- S-antigen transport-inhibiting oligonucleotide polymers
-
- T
-
- triangulation
-
- TAP
-
- Tip-associated protein
-
- TGN
-
- trans-Golgi network
-
- TM
-
- transmembrane
-
- UPR
-
- unfolded protein response
INTRODUCTION
Hepatitis B virus (HBV) infection remains a major public healthcare challenge globally. About 2 billion individuals have been infected, representing approximately 30% of world's population, and more than 257 million remain chronically infected and have a high risk to develop liver inflammation, cirrhosis, and hepatocellular carcinoma. Despite the availability and usage of an efficient prophylactic vaccine, HBV infections continue to be an important health issue, as current therapeutics, like pegylated-interferon-α and nucleos(t)ide analogues, cannot cure chronic infections. Barriers to a functional HBV cure include the persistent reservoirs for HBV replication and antigen production, the high viral burden and the impaired host innate and adaptive immune responses against HBV (Ligat et al., 2021; Nguyen et al., 2020; Revill et al., 2019; Roca Suarez et al., 2021).
HBV is an enveloped DNA virus that exclusively infects hepatocytes of humans and some non-human primates and replicates by a multifaceted process of reverse transcription. The viral life cycle requires cellular receptors as well as cellular microenvironment suitable for viral entry, uncoating, genome repair, gene expression, replication, assembly, envelopment, and ultimately virus egress from the host cell. The extremely successful spread of this virus among the human population is explained by its effective transmission strategies and its manifold particle types, including virions, empty virions, empty envelopes, and naked capsids (Hu & Liu, 2017; Prange, 2012; Seitz et al., 2020). Due to its tiny genome, HBV heavily depends on cellular machineries to thrive in infected hepatocytes. To enter, traverse and exit the cell, HBV exploits host membrane trafficking pathways. This review focuses on the present state of knowledge of intracellular trafficking routes and host factors hijacked by HBV in order to release its progeny particle types. The itineraries accompanying HBV entry and uptake, uncoating and genome delivery to the cell nucleus will not be discussed here in detail. Here, the reader is recommended to excellent reviews drafted by Blondot et al. (2016), Chuang et al. (2022), and Tian et al. (2021).
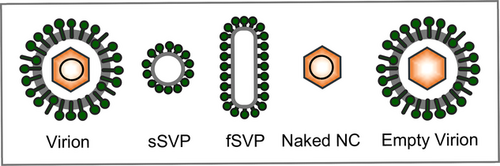
With about 3 kb in size, the partially double-stranded and relaxed circular (rc) DNA genome of HBV is one of the smallest viral genomes known. Hence, it exhibits an extremely compact organization and contains four overlapping open reading frames (ORFs), encoding the polymerase/reverse transcriptase (RT), the capsid-forming core protein and its related secretory precore protein, three related envelope proteins and the regulatory X protein. HBV attachment to the hepatocyte surface involves heparan sulfate proteoglycans (Schulze et al., 2007) and the liver-specific sodium-taurocholate cotransporting protein (NTCP), a transmembrane protein that normally mediates hepatic bile acid transport (Yan et al., 2012). The epidermal growth factor receptor is an additional factor contributing to NTCP-directed uptake of HBV (Iwamoto et al., 2019). HBV entry is considered to occur via endocytosis, rendering fusion of the viral envelope with internal membranes (Pérez-Vargas et al., 2021; Rodríguez-Crespo et al., 1995). Whether the internalization of HBV is dependent on caveola or clathrin is still a subject of debates, although clathrin is considered the favorite (Herrscher et al., 2020; Huang et al., 2012; Macovei et al., 2010). After entry, the HBV nucleocapsid (NC) is released into the cytoplasm and transported to the nucleus where the rcDNA is converted into an episomal covalently-closed-circular (ccc) DNA minichromosome which serves as a template for HBV RNA transcription (Nassal, 2015). The HBV pregenomic (pg) RNA is then incorporated in the newly formed NCs where it is retrotranscribed in the corresponding rcDNA. The cccDNA with chromosomal features is a key obstacle for a cure of chronic hepatitis B (CHB). Current approved anti-HBV drugs like therapeutic nucleos(t)ide analogs can potently block reverse transcription of the pgRNA and thus virion production, but not viral transcription and viral antigen expression (Ligat et al., 2021; Revill et al., 2019). The rcDNA-containing NC is either enveloped and released as new virions, termed the Dane particle, or redirected into the nucleus to replenish the cccDNA pool (Nassal, 2015). Besides, the newly synthesized envelope proteins can self-assemble into non-infectious, spherical and filamentous subviral particles (sSVP and fSVP, respectively) which are released into the serum of patients and even outnumber infectious virions (Figure 1) (Prange, 2012; Seitz et al., 2020).
THE HBV PRECORE PROTEIN
The HBV core gene encodes two different proteins: core, which forms the viral nucleocapsid, and precore, which is the precursor of the hepatitis B e antigen (HBeAg), a non-particulate protein, circulating in the blood of infected patients apart from the HBV particle types (Gerlich et al., 2020; Tsai & Ou, 2021). Precore has features of pre-pro-proteins and is translated with a 29-aa N-terminal sequence that directs this 25-kDa precursor protein (p25) into the endoplasmic reticulum (ER), where the signal peptide comprising the first 19 aa is cleaved off by the cellular signal peptidase, thereby generating the 22-kDa pro-protein (p22) (Figure 2a) (Tsai & Ou, 2021). The remaining 10-aa extension at the N-terminus of precore plays an important role, as it mediates the formation of an intrasubunit disulphide bond between Cys at position-7 within the pro-peptide sequence and Cys at position 61 which holds precore in a dimeric state and prevents its multimerization (Liu et al., 2021; Nassal & Rieger, 1993). P22 is next transported out of the hepatocyte via the constitutive pathway of secretion during which it is proteolytically cleaved again. Its C-terminal arginine-rich domain (ARD) carries cleavage sites for a furin-like protease, a pro-protein convertase localized in the trans-Golgi network (TGN) that cleaves p22 at Arg-154 and/or Arg-167 residues to generate the mature, secretory 17-kDa HBeAg (p17) (Figure 2b) (Messageot et al., 2003).
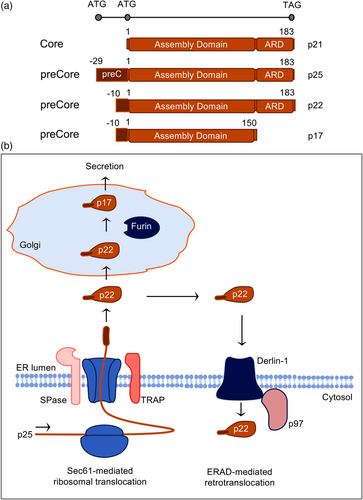
Follow-up studies analyzing precore trafficking routes disclosed entirely unexpected abilities of this protein. In pioneer works already done 35 years ago, Garcia et al. (1988) surprisingly observed that a large portion of the precore proteins, whose signal peptide had been cleaved off, recycles back to the cytoplasm rather than becoming secreted. The authors concluded that precore translocation was aborted by a yet unknown mechanism. At that time retrograde transport processes, that is, transport out of the ER back to the cytoplasm, were less known. This changes with the discovery of the endoplasmic reticulum associated degradation pathway (ERAD) which came up as a complete surprise (Sun & Brodsky, 2019). This process supports the quality control of protein folding inside the ER. It includes chaperone-mediated assistance in protein folding and the selective targeting of misfolded species which are then retrotranslocated back to the cytoplasm where they undergo ubiquitination in order to end up in degradation. To unravel the aborted translocation of precore, Duriez et al. (2008) investigated the potential involvement of the cellular ERAD machine. By using dominant-negative (DN) mutants of derlin-1 and the p97 ATPase, two major players of the ERAD pathway in mammalian cells, the cytoplasmic pool of the signal-cleaved precore disappeared. However, unlike typical ERAD substrates, retrotranslocated precore escapes ubiquitination and subsequent proteasomal degradation likely as a consequence of its low lysine content (Figure 2b).
To obtain further hints into the translocational behavior of precore, Zábranská et al. (2022) recently performed a proteomic mass spectrometry analysis of ER-entrapped precore. This led to the identification of TRAP, a protein complex consisting of four subunits (α, β, γ, δ), that firmly associates with the protein-conducting Sec61 translocon. Unlike Sec61, TRAP is an auxiliary complex, needed only for a subset of precursor proteins which together carry weak signal sequences characterized by a high glycine plus proline content (Nguyen et al., 2018) Pulldown experiments confirmed that precore interacted with all four TRAP subunits in a productive manner, as siRNA-mediated silencing of TRAP inhibited the biogenesis and secretion of p17 (Zábranská et al., 2022). The authors concluded that the signal peptide of precore is not strong enough to open the translocon channel alone, but rather requires the assistance of TRAP for translocon gating (Figure 2b).
As implied by its shortcut “e,extra,” manifold studies had shown that HBeAg does not function as a structural protein required for viral infection, replication, or assembly (Tong et al., 1991; Tsai & Ou, 2021). Rather, precore is a viral surplus protein that plays important regulatory functions, as signified by its conservation among the hepadnavirus family. The mature, secreted p17/HBeAg performs immunomodulatory functions through multiple pathways thereby facilitating the establishment of a persistent infection (Kramvis et al., 2018; Tsai & Ou, 2021). Besides, HBeAg is a tolerogen and can induce profound tolerance particularly during perinatal mother-to-child transmission of HBV (Milich et al., 1990). Cytoplasmic p22 precore protein, derived from aborted or retrograde ER translocation, had been shown to regulate innate immune and cancer signaling pathways, like, for example, suppressing the Toll-like receptor signaling pathways (Tsai & Ou, 2021). Moreover, the reducing environment of the cytoplasm can trigger a dramatic conformational switch in quaternary structure of the reimported precore dimers in such that they are now competent to co-assemble with core protein dimers into mixed heterocapsids devoid of viral replication complexes (DiMattia et al., 2013; Duriez et al., 2008). With regard to the emerging evidence of empty, genome-free HBV particles, these heterocapsids may be eventually enveloped and released thereby acting as decoys to exhaust the host immune system (Hu & Liu, 2017; Luckenbaugh et al., 2015). As will be outlined below, the cytoplasmic precore is also the fine tuner of the Rab7-dependent regulation of HBV secretion (Inoue et al., 2015). Finally, cytoplasmic p22 can enter the nucleus, which is not surprising with regard to the nuclear localization signals (NLS) shared by precore and core located within their ARD domains (Ou et al., 1989). For nuclear entry, p22 takes use of the NLS receptor karyopherin α1 (Mitra et al., 2019). As a consequence of this cross talk, the nuclear translocation of the signal transducer and activator of transcription (STAT) is out-competed as it likewise depends on karyopherin α1 (Mitra et al., 2019). The hijacking of the host nucleocytoplasmic trafficking system in turn explains how p22 is able to inhibit the antiviral signaling of interferon alpha (IFN-α). In summary, the HBV precore protein is a secreted, cytosolic, and nuclear protein with different functional roles in different subcellular locales (Zlotnick et al., 2015).
THE HBV CORE PROTEIN
The HBV core protein, also known as HBV core antigen (HBcAg), consists of 183 or 185 amino acids, depending on the genotype. It contains three distinct domains: the alpha-helical rich N-terminal domain (aa 1–140) involved in nucleocapsid assembly, a linker region (aa 141–149), and the C-terminal ARD (aa 150–181/183; also known as CTD) required for viral genome packaging and replication (Figure 2a) (Lewellyn & Loeb, 2011; Niklasch et al., 2021; Zlotnick et al., 2015). Notably, core is not only the building block of the HBV capsid/NC, but contributes to many steps of the viral life cycle including reverse transcription, genomic replication, cccDNA stability, and cellular trafficking (Chuang et al., 2022; Niklasch et al., 2021). The versatile nature of core is primarily a consequence of its dynamic phosphorylation and dephosphorylation occurring at its ARD. Several cell kinases, such as serine/arginine-rich protein kinase 1 and 2 (SRPK1, SRPK2), cyclin-dependent kinase 2 (CDK2), PKC, and polo-like kinase 1 (PLK1) have been described to be involved in core phosphorylation (Diab et al., 2017; Heger-Stevic et al., 2018; Kann & Gerlich, 1994; Ludgate et al., 2012; Wittkop et al., 2010), whereas dynamic dephosphorylation is guided by the cellular protein phosphatases PP1 and PP2A (Hu et al., 2020; Xi et al., 2021).
After HBV entry and disassembly, the incoming NCs are released into the cytoplasm. In this locale, the NC unfolds in such that its previously hidden ARD harboring NLSs flips to the capsid exterior. This conformational change facilitates NC trafficking in the cytoplasm, as the exposed NLSs can now be recognized by the cellular karyopherins importin α and β which mediate transport, along with microtubules (Chen et al., 2016; Osseman et al., 2018), to the nuclear pore complex (NPC) (Figure 3a) (Blondot et al., 2016; Schmitz et al., 2010). Notably, the external NLS exposure requires NC phosphorylation. In order to cross the NPC, the NC next interacts with nucleoporin 153 (Nup153), an intranuclear protein located at the nuclear basket (Figure 3a) (Schmitz et al., 2010). Within the nucleus, the capsid shell disintegrates and releases the viral genome into the nucleoplasm. In a multi-step process involving host DNA replication and repair factors the rcDNA is then converted into the episomal cccDNA form (Figure 3a). For an in-depth discussion of cccDNA biology the reader is referred to recent reviews from Bustamante-Jaramillo et al. (2022) and Nassal (2015).
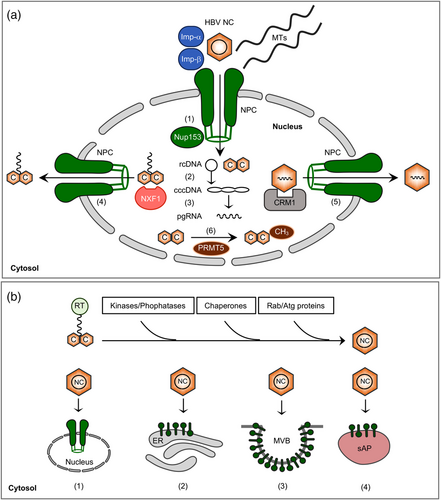
Noteworthy, the core protein does not remain in the nucleus, but is rather reexported to the cytoplasm. Studies aimed at analyzing core trafficking revealed that core shuttles continuously between the nuclear and cytoplasmic compartments (Blondot et al., 2016). This compartmentalization of core even correlates with the clinic, as core tends to be more localized to the cytoplasm in patients with chronic active liver disease, and more localized to the nucleus when the disease is mild and asymptomatic (Chu et al., 1995; Hsu et al., 1987). For nuclear export, core appears to utilize two cellular machineries. The first one involves the nuclear RNA export factor 1, also known as NXF1 or TAP (Tip-associated protein/nuclear export factor-1), that recognizes and interacts with basic, arginine-rich nuclear export signals (NES) within the ARD of core (Figure 3a) (Li et al., 2010). This finding comes up as a surprise, as it shows that core usurps the pathway of cellular mRNA export. The other export pathway depends on the chromosome region maintenance 1 (CRM1) exportin which is a major export receptor for nuclear proteins containing NESs with clustered hydrophobic rather than basic residues (Figure 3a). Indeed, Shih et al. recently succeeded (Yang et al., 2022) to discover two CRM1-dependent NESs located within the spike region of core. Interestingly, the CRM1-mediated export of core increases parallel to its nuclear concentration, arguing for transport systems that are responsive to the amount of core in a cell (Nair & Zlotnick, 2021). One potent controller of core cell trafficking and function is the protein arginine methyltransferase 5 (PRMT5) that interacts with the core protein and dimethylates arginine residues within the ARD (Figure 3a). Overexpression of PRMT5 led to increased nuclear accumulation of core, and vice versa, down-regulation of PRMT5 resulted in reduced levels of core in nuclei (Lubyova et al., 2017).
For progeny virus formation, newly synthesized core monomers assemble into T-shaped homodimers within the cytoplasm and three homodimers interact to form a trimer of dimers. The mature capsid shell is composed of either 180 or 240 core subunits with triangulation (T) numbers of 3 or 4, respectively. Both capsid species are found in infected cells, but the T = 4 capsids seem to be preferentially incorporated into virions (Roseman et al., 2005). Accordingly, the T = 3 capsids have been considered to represent dead-end products caused by aberrant assembly (Conway et al., 1997). As evidenced from in vitro studies, capsid assembly is a fine-tuned allosteric process whose kinetic strictly depends on core dimers concentration, ionic strength, and temperature (Ceres & Zlotnick, 2002; Niklasch et al., 2021; Zlotnick et al., 2015). However, in vivo, capsid assembly faces the challenge of specific packaging of the viral pgRNA together with the covalently bound RT protein which strictly requires the phosphorylation of core. Besides the host candidate kinases mentioned above, cellular proteins, such as the components of the Hsp90 chaperone complex, Hsp90, Hsp70, Hsp40, and p23, appear to facilitate the pgRNA encapsidation reaction (Figure 3b) (Hu & Seeger, 1996; Hu et al., 1997; Seo et al., 2018; Sohn et al., 2006). Meanwhile, the subsequent dephosphorylation of core, triggered by pgRNA packaging and host phosphatases, is essential for the synthesis of the minus-strand DNA catalyzed by the RT domain of the viral polymerase (Basagoudanavar et al., 2007; Hu et al., 2020; Xi et al., 2021). Simultaneously, the RNA of the hybrid is degraded by the RNaseH domain of RT. To summarize, the posttranslational modification status of core is a key trigger in the viral life cycle. A recent report provides evidence for a further post-phosphorylation regulation in such that the host peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (Pin1) binds and stabilizes the core protein in a phosphorylation-dependent manner, thereby promoting the efficient viral propagation (Nishi et al., 2020).
Since HBV capsid assembly can be recapitulated in cell-free systems and bacteria, it had largely been viewed as a spontaneously occurring self-assembly process, only regulated by the threshold levels of core proteins/dimers (Zlotnick et al., 2015). However, the crowded cytoplasm of the hepatocyte, may be an unfavorable environment for homotypic core interactions (Lingappa et al., 2005). HBV appears to face this problem by directing capsid assembly to membranes in order to raise the local concentration of core dimers. Intrinsic membrane targeting signals were first found in the duck HBV core (Mabit & Schaller, 2000) and later confirmed in the HBV core in which they had been mapped within the ARD (Bartusch et al., 2017). Mechanistically, the cellular Rab33B GTPase was identified as crucial factor for the membrane association of the HBV core protein and for the assembly and stability of capsids and their trafficking to budding sites (Bartusch et al., 2017; Doring & Prange, 2015).
Previous studies with HBV mutants as well as cell-free binding assays led to the mapping of domains of the envelope and core proteins required for capsid envelopment. Together, these studies showed that virion morphogenesis depend on the regulated interaction of rcDNA containing NCs with the viral envelope, composed of the large L, middle M, and small S protein. In particular, the envelope/NC interaction involves the L-specific matrix domain (MD) (Bruss & Thomssen, 1994), motifs in the first cytosolic loop of the S domain of S/M/L (Blanchet & Sureau, 2006; Loffler-Mary et al., 2000), the core spike tip (Böttcher et al., 1998), and/or base (Ponsel & Bruss, 2003), including a hydrophobic pocket (Pro-5, Leu-60, Leu-95, Lys-96, and Ile-97) at the core dimer interface (Le Pogam & Shih, 2002). An intriguing and long-lasting observation in hepadnaviral morphogenesis is that capsids with immature genomes are rarely enveloped (Gerelsaikhan et al., 1996; Summers & Mason, 1982). This led as early as 1982 to the “maturation signal” hypothesis postulating that a regulated conformation switch in the capsid indicates readiness for envelopment (Summers & Mason, 1982). Such a conformational signaling mechanism may be triggered by the capsid's internal genome status, capsid phosphorylation, capsid icosahedral symmetry, or others (Roseman et al., 2005). De facto, however, subsequent studies failed to convincingly trace maturation-associated structural changes of the HBV capsid. Rather, a new study questions the concept on a maturation signal driven by rcDNA formation, as it shows an intimate core-envelope cross talk in transfected hepatocytes even in the absence of viral genomes (Pastor et al., 2019).
Moreover, the maturation signal model was challenged by the observation of an excess of genome-free Dane particles both in patients and in cell culture. This observation led to the “single strand blocking” hypothesis, suggesting that capsids with an immature single-stranded genome display a blocking signal that prevents envelopment (Ning et al., 2011). Very recently, this again was called into question. By using solid-state nuclear magnetic resonance, Lecoq et al. (2021) provided new evidence for the existence of two alternate capsid conformations that could generate such maturation or blocking signals. These alternate conformations can be switched by binding a pocket factor to the hydrophobic pocket in the center of the core spikes. The natural pocket factor, either cellular or viral, is unknown thus far, but can be mimicked by the detergent Triton-X 100, signaling readiness for envelopment (Lecoq et al., 2021). Conversely, however, the team from Makbul et al. (2021) used electron cryo-microscopy and image processing and compared HBV recombinant capsids from detergent-free purifications with those purified in the same way with added Triton-X 100. The authors confirmed that Triton-X 100 binds to the hydrophobic pocket thereby inducing conformational changes of the capsid. However, since similar structural changes occur in core mutants with defects in virus assembly and secretion, it was concluded that binding of Triton-X 100 is unlikely to mimic structural maturation (Makbul et al., 2021). Rather, instead of the existence of a maturation or blocking signal, different virion secretion phenotypes may simply be explained by the speed of trafficking and envelopment. In this view, a recent study uncovered that the linker region of the viral capsid can modulate the timing of capsid-envelope interactions in an HBV genotype-dependent manner and consequently the speed of virion maturation and release (Xi et al., 2022).
As an alternative option to NC envelopment and secretion, the newly produced NCs, like those from de novo infection, may recycle their rcDNA to the nucleus to expand or replenish the cccDNA pool (Figure 3b) (Tuttleman et al., 1986). The fact that HBV mature progeny NCs can disassemble without leaving the host cells is a very unique property of hepadnaviruses. Notably, NC disassembly is an essential step for HBV infection as well as for cccDNA amplification. However, it is less known whether uncoating of HBV rcDNA from virion-derived NCs during de novo infection and the cytoplasmic progeny mature NCs during cccDNA amplification may take place at different subcellular compartments and possibly via distinct mechanisms. In favor of different routes are recent observations of distinct usage of host DNA polymerases, as the de novo cccDNA synthesis requires cellular DNA polymerase κ and λ (Qi et al., 2016), while the amplification of cccDNA relies on DNA polymerases α (Tang et al., 2019). This led to the suggestion that the two cccDNA synthesis pathways might be fueled and imported into the distinct subdomains of the nucleus, possibly governed by host factors.
Irrespective of their final destination, either leading to recognition by the envelope proteins or their fueling into the recycling pathway, cytoplasmic capsid trafficking depends on microtubules (MT) along with the dynein light chain LL1 as a functional interaction partner linking capsids to the dynein motor complex (Osseman et al., 2018). In support, a follow-up study, probing the spatiotemporal patterns of HBV multiplication by using a microscopic approach for multiplex detection of viral nucleic acids and proteins, showed that NC transport out of perinuclear areas requires MT integrity (Yue et al., 2021). Mechanistically, the HBV X protein has been shown to up-regulate the microtubule-associated protein 1S (MAP1S) thereby promoting stable MT formation implicating that HBV manipulates the cytoskeleton to facilitate its own transportation (Guan et al., 2021).
THE HBV ENVELOPE PROTEINS
The HBV envelope is composed of cellular lipids and the three related glycoproteins, termed S, M, and L protein. Despite their relatedness, they differ in function, as S forms the scaffold of the envelope, while L is required for infectivity and NC envelopment. Curiously, no essential function could be assigned to the M protein thus far (Bruss & Ganem, 1991; Le Seyec et al., 1998). Similar to the precore/core ORF, the three envelope proteins provide a further striking example of hepadnaviral economy, as they are expressed from one single open reading frame by the use of three different start codons and a common stop codon. Hence, all three proteins share the 226-aa sequence of S, that is, the S domain. M is N-terminally extended by the preS2 domain of 55 aa, and L encompasses in addition the preS1 domain of 108, 118, or 119 aa, depending on the genotype. For simplification, all preS1 coordinates provided herein have been converted to the 108 aa version of HBV, genotype D (Figure 4a). All three proteins are cotranslationally integrated into the ER membrane, directed by the action of an uncleaved signal-anchor and a stop-transfer sequence encoded within the first and second transmembrane (TM) segments (TM1, TM2), respectively, of their S domains (Prange, 2012; Seitz et al., 2020). Two further membrane-spanning segments are predicted in the C-terminal third of the S domain. The amphipathic TM1 and the hydrophobic TM2 are separated by the hydrophilic cytosolic loop I (CYL-I), while TM2 is followed by a hydrophilic ectodomain, termed the “a”-determinant or antigenic loop (AGL), that contains the major conformational epitope of the envelope and a N-glycosylation site (Asn-146) (Figure 4a). Both hydrophilic loops contain several cysteines, involved in intra- and intermolecular disulfide bonding thereby creating an oligomeric network in the viral membrane (Mangold & Streeck, 1993). The topology of the M protein is highly similar to that of S and its hydrophilic preS2 domain is translocated into the ER membrane by the TM1 and TM2 signals of the S domain. The M-specific preS2 region is N-glycosylated at Asn-4. Beside N-glycosylation, the M glycoprotein of HBV genotype D is also O-glycosylated at Thr-37 site in its preS2 domain (Schmitt et al., 1999; Werr & Prange, 1998).
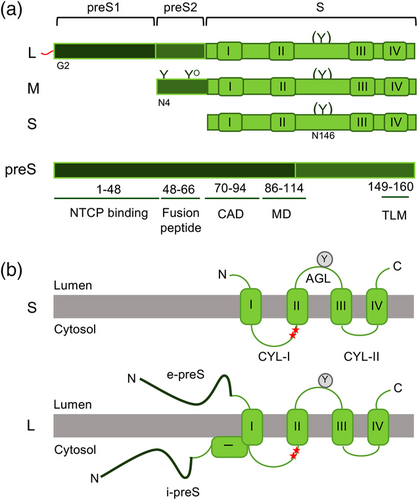
The L protein is the queen among the HBV envelope proteins and has therefore achieved remarkable attention. Owing to its tiny genome, HBV has evolved by packing a maximum of information in a minimum of polypeptide sequence, as exemplified by the preS (preS1 plus preS2) region of L. The preS part of L plays an essential role in a multiplicity of interactions with viral and cellular proteins (Figure 4a). The N-terminal 77 aa are the major determinant of infectivity, as they mediate viral entry into the host cell. The binding to the NTCP receptor is essentially based to the first 48 aa and additionally requires the N-terminal myristyl moiety attached to Gly-2 of L (Persing et al., 1987; Schulze et al., 2010; Yan et al., 2012). Further down the preS sequence harbors hydrophobic stretches that are concentrated between residues 50–70 and may mediate membrane interactions. In support, a very recent proposal locates a fusion peptide, involved in HBV entry, exactly to this hydrophobic region (Pérez-Vargas et al., 2021). A third vital region is localized at the preS1/preS2 border, including residues between 92 and 113, that comprise the MD region (Bruss & Thomssen, 1994). This region establishes contacts to the nucleocapsid and is a key element for nucleocapsid envelopment. In addition, NMR and X-ray crystallography analyses of preS1 led to the identification of several motifs that mediates binding of L to γ2-adaptin, a member of the cellular clathrin adaptor protein (AP) family which mediates protein sorting in endocytic and secretory transport pathways (Jürgens et al., 2013). The L-γ2-adaptin interaction contributes to virion export (Rost et al., 2006) and mimics membrane-trafficking motifs of host proteins (Jürgens et al., 2013). Finally, the stretch of aa 70–94 of preS1is involved in the interaction with the cognate heat shock protein Hsc70 that determines the cytosolic anchorage of preS during cotranslational ER integration and in such regulates the extraordinary topology of L (Lambert & Prange, 2003; Loffler-Mary et al., 2000). A cell-permeable peptide was identified between the aa 41 and 52 of preS2 whose amphipatic α-helix mediates cell access by acting as a translocation motif (Figure 4a) (Oess & Hildt, 2000). A very recent NMR study of preS reveals novel sites for phosphorylation (Fogeron et al., 2021). Intriguingly, all identified phosphorylation acceptor sites are located in or just next to regions closely linked to the different preS functions pointing to a possible role thereof in the regulation of preS function.
The multifunctional nature of L remained a mystery for a long time. In particular, it was less comprehensible how preS could operate at the virion outside during receptor binding and in the virion inside during NC envelopment. This puzzling was unraveled in three independent studies in the middle of the nineties. Without going into experimental details, these studies enlightened a dual transmembrane topology of L, enabling it to dispose its N-terminal preS domain to both the cytosolic (i-preS) and luminal (e-preS) side of membranes (Bruss et al., 1994; Ostapchuk et al., 1994; Prange & Streeck, 1995). Like its S and M relatives, L is cotranslationally translocated into the ER membrane by the proximal topogenic signals of its S domain concomitant with N-glycosylation occurring at Asn-146 (Eble et al., 1986; Heermann et al., 1984). During this process, its preS domain fails to be translocated and initially remains cytosolic. Thereafter, approximately half of the L molecules posttranslationally translocate the preS region across the membrane (Figure 4b). Due to its posttranslational translocation preS failed to be modified at its N-glycan acceptor sites at Asn-4 within preS1 and Asn-4 within preS2 (Bruss et al., 1994; Ostapchuk et al., 1994; Prange & Streeck, 1995). Mutational profiling of preS discovered its aa 70–94 as a “cytosolic anchorage determinant (CAD)” responsible for cotranslational preS retention, as its deletion supported the emergence of double- and triple-glycosylated forms of L at the ER, modified within preS (Loffler-Mary et al., 2000). The function of the CAD relies on its productive interaction with Hsc70 in conjunction with Hsp40(Lambert & Prange, 2003; Loffler-Mary et al., 2000), whereas the ER luminal BiP chaperone interacts with e-preS likely to provide the driving force to pull on the preS chains into the ER lumen, until L has adopted its final dual topology (Awe et al., 2008). Altogether, the early studies advocated that the dual topology of L is established already at the ER membrane before envelopment of NCs. However, more than 20 years later, a follow-up, cryo–electron microscopy study indicates that the topological switch of L also takes place in extracellular virions as a distinct maturation step ensuring the extraordinary transmission efficiency of HBV (Seitz et al., 2016).
HBV AND THE COPII MACHINERY
The envelope proteins are exported out of the ER in a seemingly sophisticated manner by co-opting the coat protein complex II (COPII) trafficking pathway. Discovered in the 1990s, COPII has been identified as a vesicular transport machinery, moving secretory cargos from the ER to the Golgi apparatus via two coexisting mechanisms. The first one is passive, termed bulk flow, in which any ER-localized cargo can be non-selectively encapsulated into COPII vesicles. The second, more efficient mechanism is an active sorting of cargo proteins based on specific signals (Barlowe & Helenius, 2016; Khoriaty et al., 2012).
COPII assembly starts with the activation of the small GTPase Sar1 and its recruitment to the ER with assistance of its membrane-bound guanine nucleotide exchange factor Sec12. Upon GTP-loading, activated Sar1 next recruits the Sec23/Sec24 dimer to build the inner coat layer. Sec24 serves as a cargo adaptor of the COPII machinery and specifically binds to cargo proteins via distinct binding motifs, whereas Sec23 modulates the GTP cycle of Sar1. After this, the tetrameric Sec13/Sec31 complex gets recruited to the Sar1–Sec23/24 complex in order to form the outer coat layer, sustain the membrane curvature and promote vesicle budding (Figure 5) (Barlowe & Helenius, 2016; Khoriaty et al., 2012; Wendeler et al., 2007). A split-ubiquitin based yeast two hybrid assay using the HBV L and S proteins as baits led to the identification of Sec24A that interacted with the S domain Arg-78/Arg-79 dipeptide signal located at end of CYL-1 (Zeyen et al., 2020). Mammalian cells contain four Sec24 paralogs that can be divided into two subclasses, Sec24A/B and Sec24C/D, sharing about 60% sequence identity within the subclasses (Wendele et al., 2007). Quite unusual, the HBV envelope turns out to be a very specific client of the Sec24A isoform together with Sec23B whose functions could not be substituted by their related paralogs. SiRNA-mediated silencing of Sec24A and Sec23B as well as of Sar1 and Sec31 blocked ER export indicating that HBV uses COPII for trafficking (Figure 4) (Zeyen et al., 2020). Although these pilot studies were conducted with the HBV genotype D strain, the dibasic Arg-78/Arg-79 motif likely resembles a bona fide ER export code as it is conserved in all 10 HBV genotypes.
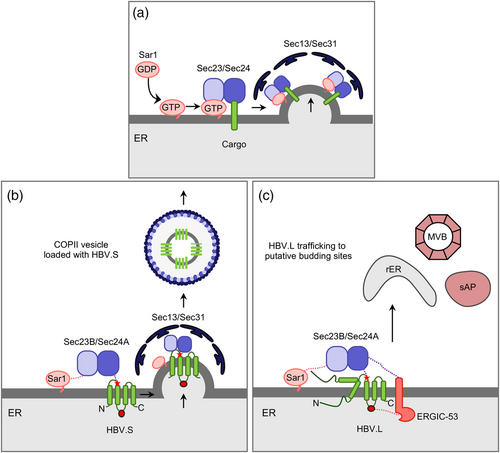
HBV AND THE GLYCOSYLATION MACHINERY
Glycosylation is one of the most common posttranslational modification of viral proteins and plays important biological roles, like protein folding, virus attachment to host cell receptors, and inhibition of antibody neutralization. N-linked glycosylation occurs on asparagine residues of Asn-X-Ser/Thr sequons, where X is any amino acid except proline, and is catalyzed by the ER-resident oligosaccharyltransferase (OST) (Dobrica et al., 2020). Glycoprotein maturation in the ER requires the stepwise removal of the two terminal glucose residues of the N-linked glycan by ER α-glucosidases I and II. Trimming of these terminal glucose residues provides glycoprotein substrates for calnexin/calreticulin-assisted folding. These two ER-resident lectins specifically interact with mono-glucosylated glycans attached to glycoprotein intermediates, thereby preventing potential aggregation. Removal of the last glucose unit releases glycoproteins from the calnexin/calreticulin cycle (Dobrica et al., 2020). Indeed, interaction with calnexin was demonstrated for both, the HBV M and L proteins (Werr & Prange, 1998; Xu et al., 1997). Next, other enzymes like mannosidases, glucosidases, sialyl-, fucosyl-, or galactosyl-transferases located at the ER-Golgi apparatus further process the glycoproteins thereby generating three types of N-glycans based on the structures, including oligomannose, hybrid, and complex-type N-glycan structures.
Although the HBV envelope proteins are not heavily glycosylated, they exploit the host N-glycosylation pathway in a very peculiar manner. All three proteins are modified at Asn-146 in the AGL region (Heermann et al., 1984). Strikingly, this sequon is modified only in half of the envelope proteins resulting in similar amounts of glycosylated and non-glycosylated isoforms of S, M, and L. The partial N-glycosylation is quite uncommon and has been assigned to the presence of a cysteine residue in the Asn-X-Ser/Thr sequon. The Cys-145 residue is involved in intra- and intermolecular disulfide bridge formation (Mangold & Streeck, 1993; Mangold et al., 1997) and in such may hamper the complete access for the cellular OST enzyme. Another N-glycosylation site is located at Asn-4 of the preS2 domain that is always occupied in M but never in L due to their different features in ER translocation (Prange, 2012).
Interestingly, the HBV viral and subviral envelopes differ in their needs of N-glycans added to Asn-146. Several studies reported that both N-linked glycosylation and N-linked glycan processing at Asn-146 is essential for the release of viral particles while the secretion of sSVP preceded normally in the absence of added sugar moieties (Dobrica et al., 2020; Hassemer et al., 2017; Julithe et al., 2014; Lu et al., 1995; Mehta et al., 1997). One explanation for this puzzling finding is the recent discovery of the host protein ERGIC-53(ER-Golgi intermediate compartment [ERGIC] 53 protein; also known as lectin mannose-binding 1 protein, LMAN-1) as a selective interaction partner of HBV (Zeyen et al., 2020). ERGIC-53 is a high mannose-specific lectin that cycles cargoes between the ER and the cis-Golgi through COPII- and COPI-dependent pathways. It is a type I integral membrane protein with a C-terminal transmembrane domain and a luminal cargo recognition domain interacting with N-linked glycans of cargo proteins (Appenzeller et al., 1999). The silencing of ERGIC-53 in HBV-replicating liver cell cultures inhibited HBV virion egress without affecting HBV sSVP release (Zeyen et al., 2020), implicating that it may serve as a device to segregate intracellular trafficking routes of HBV particle types (Figure 5). Mutational analysis provided evidence that the L protein interacts with ERGIC-53 in an Asn-146-glycan-dependent manner (Zeyen et al., 2020). Thus far, only an amazing small number of cellular glycoproteins is trafficked in an ERGIC-53-dependent manner (Zhang et al., 2009). To account for this, interaction analyses revealed that the lectin ERGIC-53 not only recognizes sugars but also combined oligosaccharide/peptide structures. A carbohydrate- plus conformation-dependent recognition by ERGIC-53 may likewise apply to the HBV envelope types. Despite sharing the same primary sequences, the configuration of the L-specific S domain within the viral envelope may differ from the S protein within the subviral envelope thereby warranting that ERGIC-53 can segregate the maturation pathways of the HBV viral and subviral particles.
More recently, the N-glycosylation of HBV has been identified as a target of restriction imposed by the cellular serine incorporator 5 (SERINC5) that normally guides lipid biosynthesis and transport. Originally, the multipass transmembrane protein SERINC5 was shown to be incorporated into budding human immunodeficiency virus type 1 (HIV-1) particles thereby reducing their infectivity which in turn is counteracted by the HIV-1 Nef protein (Rosa et al., 2015; Usami et al., 2015). In case of HBV, ectopic overexpression of SERINC5 restricted HBV progeny production and release while its silencing had opposite effects (Liu et al., 2020). Mechanistically, the antiviral mode of action of SERINC5 relied on a block of the HBV-specific N-glycosylation thereby impeding the proper trafficking and export of viral particles (Liu et al., 2020). However, it is yet unclear how SERINC5 inhibits Asn146-linked glycosylation of the HBV envelope.
HBV AND THE AUTOPHAGY MACHINERY
Xenophagy is a form of selective autophagy that targets invading pathogens for lysosomal degradation, thus, operating as an innate immune response at the cellular level (Dreux & Chisari, 2010). Probably as a counter defense, HBV have evolved strategies for hijacking the cellular autophagic machinery to support its replication and dissemination. Although multiple intersections between HBV infection and the different stages of autophagy have been reported to occur in cultured cells, during natural infection and in transgenic mice (Doring et al., 2018; Li et al., 2011; Sir et al., 2010; Tian et al., 2011), the underlying mechanisms are not fully elucidated. For example, studies have implicated either the viral X protein or the viral S protein as a trigger for autophagy induction (Li et al., 2011; Sir et al., 2010). In case of S, it can trigger the ER stress-induced unfolded protein response (UPR) and activate inositol-requiring protein-1α (IRE1α) and activating transcription factor 6(ATF6) signaling pathways to induce UPR-related autophagy (Li et al., 2011). This helps the virus to favor its production, as the NC assembly reaction critically requires the autophagy elongation complex Atg5-Atg12-Atg16L1 as a physical scaffold (Doring & Prange, 2015; Doring et al., 2018). The finding of this complex is owed to an siRNA screen aimed at identifying cellular Rab GTPases guiding HBV maturation and release (Zeyen & Prange, 2018). One prominent hit of this screen was Rab33B that not only guides intra-Golgi retrograde trafficking but is also connected to the autophagy network. Rab33B is able to interact with the Atg5-Atg12-Atg16L1 complex in a GTP-dependent manner and hence uses autophagic proteins as effectors (Itoh et al., 2008). Biochemical and cell imaging studies of HBV-replicating HuH-7 cells showed that the silencing of Rab33B as well as of Atg5, Atg12, and Atg16L1 substantially impaired the synthesis, assembly and/or stability of HBV core/capsids concomitant with improper NC formation and trafficking to budding/envelopment sites (Figure 6) (Doring & Prange, 2015; Doring et al., 2018). HBV accesses the Atg5-12-16L complex via a direct interaction of its core protein with the intrinsically disordered region of Atg12. Mechanistically, deficient Rab33B reduced the membrane association of core thereby provoking aberrant core/capsid accumulations that are prone to degradation. In contrast, subsequent autophagosome maturation and closure events were unnecessary for HBV replication, as neither the lipidation of LC3B not Atg4 were required for this process (Doring et al., 2018). As explanation discussed here, HBV may use the Atg5-12/16L1 conjugate in a noncanonical manner (i.e., as a distinct function mediated by individual autophagy proteins) (Doring et al., 2018). Nonetheless, the LC3 complex can interact with the S envelope protein (Li et al., 2011), eventually to serve as a source of membranes for the viral envelopment process.
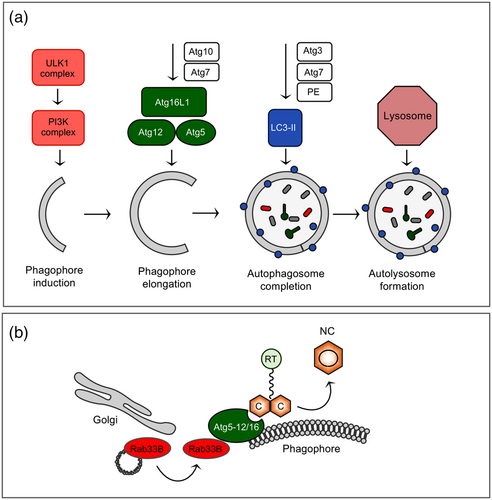
With regard to the late stages of autophagy, there is plenty of evidence that HBV does not end up in the destructive autolysosome. To escape from autophagic degradation, HBV interferes with the fusion of autophagosomes with lysosomes. This fusion process involves the Rab7 GTPase and the synaptosomal-associated protein 29 (SNAP-29) SNARE whose expression is down-regulated by the presence of HBV, likely to prevent viral clearance (Lin, Wu, Wang, Kemper, et al., 2019). Consistently, the silencing of either Rab7A or SNAP-29 increased HBV production, while their overexpression had opposite effects (Inoue et al., 2015; Lin, Wu, Wang, Kemper, et al., 2019). In addition, the HBV X protein had been shown to inhibit autophagic degradation by impairing the V-ATPase-driven acidification of lysosomes (Liu et al., 2014). With regard to Rab7A, Inuoe et al. (2015) observed that HBV even activates this GTPase despite its antiviral activity. By defining the HBV component responsible for Rab7A activation, the authors uncovered the precore protein, located in the cytoplasmic locale, to be sufficient. In order to resolve this obvious contradiction, the authors hypothesize that HBV might put a brake on its own secretion through precore-mediated Rab7A activation to minimize antiviral immune responses.
Besides Rab7A, the Rab5B GTPase is likewise involved in the late steps of the HBV infection cycle. A siRNA screen targeting 62 human Rab proteins in HepG2.2.15 cells led to the identification of Rab5B whose inactivation increased the yields and egress of progeny virions, as it is the case for Rab7A (Inoue et al., 2019). However, in difference to Rab7A, Rab5B had been suggested to control the trafficking of the L envelope protein from the secretory system to the multivesicular body (MVB), as its depletion led to an accumulation of L at the ER. Nevertheless, ER-arrested L is still competent to envelope the NCs with the assistance of the endosomal sorting complexes required for transport (ESCRT) machinery (Inoue et al., 2019), implicating that the ER as well as the MVB can support HBV budding.
HBV AND THE ESCRT MACHINERY
The ESCRT machinery is an ancient system for membrane remodeling and scission. ESCRT proteins were originally identified in budding yeasts as vital factors involved in the biogenesis of the MVB. In the course of MVB biogenesis, the ESCRT machinery is essential for the sorting of cargo proteins into intraluminal vesicles either for degradation, lysosomal functions, or exosomal release (Vietri et al., 2020). Beside this classic function, the past few years have seen an explosion of novel ESCRT functions such as membrane abscission step in cytokinesis, plasma membrane wound repair, neuron pruning, nuclear envelope reformation, autophagy, and unconventional protein secretion (Vietri et al., 2020). Importantly, viruses, as obligate intracellular parasites, have evolved to hijack the ESCRT machinery or part of it to execute their replication cycle (Votteler & Sundquist, 2013). Enveloped viruses are enwrapped by cellular membranes and usually egress from infected cells without necessarily killing their host. In seminal works, published in 2003, a surprising role for ESCRT proteins was identified in the budding step of some enveloped retroviruses, including HIV-1(Martin-Serrano et al., 2003; Strack et al., 2003; von Schwedler et al., 2003). Since this initial discovery, many other enveloped viruses, especially those budding at the plasma membrane, have been shown to utilize this pathway (Votteler & Sundquist, 2013). In principle, enveloped viruses recruit the ESCRT machinery through the function of specific peptide motifs within their structural proteins referred to as late assembly domains that can be divided in three distinct prototype classes (PT/SAP, YPXnL/LXXLF, and PPXY), even if it has recently been suggested that other motifs could act as L domains. The HBV core protein contains two candidate late assembly domain-like motifs, PPAY (aa 129–132) and PNAP (aa 135–138), what prompted the Shih´s group to start ESCRT research on HBV (Kian Chua et al., 2006).
The modular organization of the ESCRT system involves three core complexes, termed ESCRT-I, ESCRT-II, and ESCRT-III, and the terminal Vps4 ATPase. The machinery operates in a sequential manner in such that the early acting ESCRT-I and ESCRT-II capture and concentrate ubiquitinated transmembrane cargo prior to transfer to the late acting ESCRT-III and Vps4 membrane scission machinery (Figure 7). The Vps4 ATPase ensures the disassembly and recycling of the ESCRT components (Vietri et al., 2020; Votteler & Sundquist, 2013). Thus far, there are no pharmacological inhibitors available for the ESCRT pathway, but overexpression of DN mutants of Vps4 (ATP hydrolysis- or ATP binding-defective mutants) is usually effective. With these Vps4 mutant tools, Shih et al. (Kian Chua et al., 2006) demonstrated that their overexpression in HBV-replicating cell cultures inhibited HBV secretion in a dose-dependent manner. Two subsequent papers, published one year later at nearly identical days, confirmed and extended these findings in such that DN versions of Vps4 blocked HBV particle egress without affecting HBV sSVP release (Lambert et al., 2007; Watanabe et al., 2007). As would be expected, HBV budding also required scission functions provided by ESCRT-III, as evidenced by the overexpression of DN GFP-tagged ESCRT-III subunits, like the CHMP3 and CHMP4 subunits. Follow-up studies aimed at diagnosing a role for the upstream ESCRT complexes, led to the unexpected finding that HBV depends on the action of the tetrameric ESCRT-II complex, composed of EAP20, EAP30, and EAP45 (Stieler & Prange, 2014). All viruses that bud in an ESCRT-dependent manner share the requirements of ESCRT-III and Vps4, but rarely engages ESCRT-II (Votteler & Sundquist, 2013). Rather most of those viruses use ESCRT-I or the accessory Alix protein to enter the cascade. With regard to ESCRT-I, there are conflicting data arguing in favor or against its role in HBV budding. Upon RNAi-mediated silencing of ESCRT-I subunits in HBV-replicating liver cell lines, one report demonstrated that the Vps28 component of ESCRT-I is essential for HBV propagation while another study claimed its dispensability (Hoffmann et al., 2013; Stieler & Prange, 2014). Because several ESCRT factors are known to be versatile, multifunctional, with pleiotropic effects on multiple targets, some differences in the experimental conditions may impact those different outcomes. Together, HBV can be clearly classified as an ESCRT-dependent virus, although it may not require the entire ESCRT machinery.
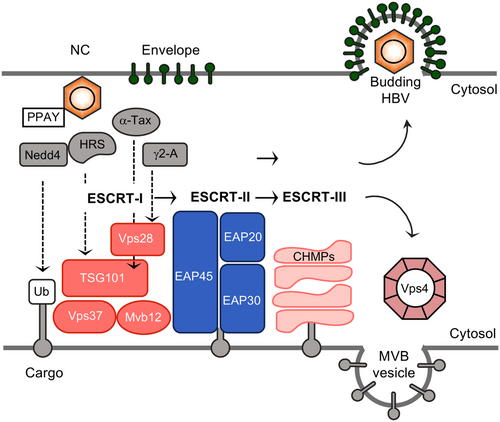
Beyond that, siRNA screening identified the ESCRT-0 complex as a critical checkpoint regulating different egress paths of HBV particle types (Chou et al., 2015). ESCRT-0 consists of two subunits, HRS (hepatocyte growth factor-regulated tyrosine kinase substrate) and STAM1/2 (signal transducing adaptor molecule1/2). ESCRT-0, in spite of its name, is not a core component of the membrane budding and scission machinery, but is merely required for initiating MVB-dependent cargo sorting by functioning as a targeting adaptor for the ESCRT cascade. Aberrant expression of the HRS subunit of ESCRT-0 can inhibit HBV replication, thus reducing the amounts of extracellular virions, yet increasing the release of naked capsids (Chou et al., 2015). An inverse correlation between secreted virions and secreted naked capsids was likewise observed upon functional inactivation of ESCRT-III and Vps4 (Bardens et al., 2011; Watanabe et al., 2007), demonstrating that HBV naked capsid budding does neither require ESCRT-0 nor a functional ESCRT scission machinery. This led to the speculation that the egress routes of virions and naked capsids share the common origin of precursors, and along the way, they branched out from each other. It remains to be clarified how the ablation of ESCRT activities can even stimulate naked capsid release in a pronounced manner. In regard to their membrane sealing functions (Vietri et al., 2020), the lack of ESCRT may prevent a rapid repair of plasma membrane holes through which naked capsids – with about 30 nm in diameter – may escape from cells.
In case of retroviruses, their Gag proteins – the antagonists of the HBV core protein - encode all the activities that are required for virion assembly and release including the recruitment of the ESCRT machine. Conversely, HBV budding and egress depends on both, the envelope and core structural proteins, which raises the question, how the virus gains access to ESCRT. In case of the HBV envelope, access motifs appeared to be decoded within the L protein, because the secretion of sSVPs (lacking L) is ESCRT-independent (Lambert et al., 2007; Watanabe et al., 2007), while the secretion of subviral filaments (containing L) requires ESCRT-III and Vps4 (Jiang et al., 2015). One report identified human α-taxilin as an L-specific interaction partner that is essential for the release of virions. By virtue of an L-domain like motif (YXXL, YAEL), α-taxilin can also interact with Tsg101, one subunit of ESCRT-I (Figure 6) (Hoffmann et al., 2013). These experiments had been interpreted as indication that α-taxilin may act as an adaptor to link the ESCRT machinery to the HBV envelope proteins. Another candidate adaptor protein may be γ2-adaptin that interacts exclusively with the i-preS topology of L (Figure 7) (Hartmann-Stühler & Prange, 2001; Lambert et al., 2007). γ2-Adaptin is a member of the AP family, guiding antero- and retrograde trafficking between the Golgi network and endosomes. γ2-Adaptin is highly similar to γ1-adaptin, one large subunit of the AP-1 complex, but appears to function differently, as evidenced by its unique ubiquitin binding capacity and its ability to interact with the ESCRT-I subunit Vps28 and the ESCRT-III subunit Chmp2 (Doring et al., 2010; Rost et al., 2006). The HBV capsid, in turn, may enter the ESCRT network by hijacking γ2-adaptin, the Nedd4 ubiquitin ligase, and/or the ESCRT-II complex that all interact with the core protein (Figure 7) (Rost et al., 2006; Stieler & Prange, 2014). Notably, core binds to Nedd4 partly via its late domain-like PPAY sequence, as evidenced by mutational analysis. Individual alanine substitutions of the PPAY motif inhibited HBV egress (Ponsel & Bruss, 2003). However, since these mutations also impaired proper NC assembly and envelopment (Ponsel & Bruss, 2003; Pastor et al., 2019), the late domain property of the HBV´s-specific PPAY is not yet fully clarified. Nonetheless, a productive interplay between Nedd4 and the late domain-like motif in HBV core was corroborated in elegant swapping experiments in which this peptide sequence (129PPAYRPPNAP138) was installed into the murine leukemia virus (MLV) Gag protein (Garcia et al., 2013). Thereby, the PPAY motif of core, but not its PNAP motif, demonstrated L-domain activity in the context of MLV production that could be stimulated by overexpressed Nedd4. The involvement of Nedd4 prompted the question whether core is a target for ubiquitination. However, against expectation, several independent studies failed to verify a ubiquitination of core at its Lys-96 residue (Garcia et al., 2009; Langerová et al., 2020; Rost et al., 2006) which is essential for NC envelopment and virion egress (Ponsel & Bruss, 2003). Rather, Lubyova et al. (2017) found that core ubiquitination occurs on the Lys-7 residue, which lies within the sequence YKEF, resembling the tyrosine-based trafficking motif YXXF. In general, YXXΦ motifs are involved in receptor internalization from the plasma membrane and protein targeting to lysosomes, endosomal compartments, and the trans-Golgi network (Kozik et al., 2010). While Lys-7 of core is dispensable for HBV propagation (Ponsel & Bruss, 2003), it has been suggested to promote virus trafficking and release (Lubyova et al., 2017).
HBV AND EGRESS MACHINERIES
Recent reports open up the perspective that HBV morphogenesis and egress may additionally depend on the lipoprotein secretory pathway. Apolipoprotein E (ApoE) is one key player in the cell lipid metabolism and transport and at the same time promotes early and late steps in the HBV life cycle (Tréguier et al., 2022). ApoE silencing in HBV-producing liver cells impaired the HBV envelopment reaction but was dispensable for the release of naked capsids (Qiao & Luo, 2019). As the HBV S protein can directly interact with ApoE (Gomez-Escobar et al., 2021), it likely captures ApoE in the virion envelope thereby supporting HBV infectivity. Similar dual functions in promoting virus assembly and virus infection had been described for the hepatitis C virus, a cousin of HBV (Tréguier et al., 2022).
Tetherin (also known as BST2) is yet another host protein incorporated into the envelope of HBV virions either by chance or by use. In 2008, the IFN-stimulated gene product tetherin was discovered as the host restriction factor responsible for preventing the release of HIV-1 with mutated Vpu from host cells (Neil et al., 2008; Van Damme et al., 2008). In the meantime, it becomes evident that tetherin blocks the egress of a variety of enveloped viruses through tethering the budding virions on the surface of the plasma or intracellular membranes. Using different HBV expression systems, ectopically expressed or IFN-induced tetherin strongly inhibited HBV release, but not the secretion of SVPs or naked capsids (Lv et al., 2015; Yan et al., 2015). Confocal microscopy and TEM analysis revealed that overexpressed tetherin locked HBV budding in cytoplasmic cisterna compartments reminiscent for MVBs (Yan et al., 2015). To restrict HBV egress and spread, tetherin physically interacts with the HBV envelope, in particular with the fourth TM domain of the S protein (Miyakawa et al., 2015). However, there is accumulating evidence that its anti-HBV activity is comparatively modest, implicating that HBV may antagonize tetherin. In this respect, possibilities like an HBV-induced hepatocyte-specific environment and/or tetherin inhibition through its interaction with the HBV S protein are discussed.
Since progeny viral proteins are hardly detectable at the plasma membrane of the hepatocyte, there is scientific agreement that HBV is an intracellularly budding virus. However, the nature of the budding site remains enigmatic even after its discovery 55 years ago. By inspecting review articles and illustrations on HBV biology, budding sites are either depicted at the secretory apparatus, especially at the ER, within the MVB pathway system, and/or at secretory autophagosomes (see also Figure 4).
If MVBs may participate in HBV maturation, progeny virions are likely released through a pathway that resembles exosome secretion. In favor of this concept are new findings implicating to an intimate crosstalk between the HBV L protein and CD63, a popular marker of MVBs and exosomes (Ninomiya et al., 2021). There are, however, obstacles arguing against an MVB-guided HBV exocytosis. First, HBV and its structural components are known to be degraded in lysosomes which are reached via movement through the endosomal/MVB apparatus (Inoue et al., 2015; Liu et al., 2014; Lin, Wu, Wang, Liu, et al., 2019). Therefore, HBV-specific cell imaging studies must be considered with some caution, as they may not differentiate between progeny viral particles destined for spread or destruction. Second, the N-glycan pattern of extracellular virions is very difficult to reconcile with an exclusive MVB export pathway, as it implicates a virus transit through the Golgi stacks. During glycoprotein transport, Golgi-residing enzymes modify N-linked mannose-rich oligosaccharides into complex glycans that are resistant to treatment with Endoglycosidase H (EndoH). By implication, an EndoH sensitivity of glycoproteins indicates the presence of high-mannose glycans and the absence of glycoprotein modification by Golgi-residing enzymes and vice versa. Studies aimed to decipher the N-glycan pattern of extracellular HBV virions released from HBV-producing liver cell lines revealed the resistance of the L-specific N-glycan to Endo H digestion along with Golgi-specific glycan trimming (Dobrica et al., 2020; Julithe et al., 2014; Zeyen et al., 2020), indicating that the viral envelope must have traversed the medial/trans Golgi apparatus en route to the exterior. Likewise, N-glycan profiling of patient-derived HBV particles demonstrated the presence of partially sialylated, biantennary complex-type glycans (Schmitt et al., 2004) which, in turn, argues in favor of a Golgi transit of the virus. Although it cannot be excluded, it seems absurd that viral particles budded within the MVB are transported back to the Golgi stacks. Third, the functional inactivation of the ESCRT-II complex inhibited the HBV export prior to N-glycan processing occurring within the Golgi complex (Zeyen, et al., 2020). From a spatial/temporal point of view, ESCRT would then act upstream rather than downstream of the N-glycan processing enzymes. This led to the proposal that ESCRT-driven HBV assembly and budding may (also) occur at the ER. A similar budding scenario had been described for the enveloped flavivirus that recruits a unique subset of ESCRT for its budding on the ER (Tabata et al., 2016). Together, the precise itinerary of HBV out of the cell remains to be decoded and warrants further investigation.
Despite the rapid and innumerable development of imaging technologies aimed to understand viruses, HBV is an exceptionally difficult virus candidate and poses challenges to visualize its life cycle, especially the late steps, in live cells. Firstly, HBV is particularly difficult to genetically modify because of the small genome and the overlapping ORFs. And secondly, the different particles types as well as the huge amount of subviral particles hamper to track those particles that correspond to actual infectious viruses. There are several reports attempted to label HBV by using lipophilic dyes, genetically encoded fluorescent proteins, genetically encoded tags, and click protein chemistry that are summarized in two excellent overviews (Deffieu & Gaudin, 2019; Zhang et al., 2016). Nonetheless, to address the dynamic behavior of complete infectious HBV particles, innovative strategies are needed to optimize the labeling strategies and to overcome current technical limitations.
HBV AND SUBUNIT PARTICLES
The formation and extracellular release of non-infectious spherical sSVP is a trademark of hepadnaviral infections, which was the first aspect of the viral life cycle to be discovered, but the last to be uncovered. Remarkably, sSVP, also referred to as hepatitis B surface antigen (HBsAg) particles, are secreted in extreme surplus compared with the infectious HBV particles and are suggested to act as decoys to the immune system, thereby exhausting B and T cell responses and contributing to viral persistence (Mohebbi et al., 2018). In the clinic, the loss of HBsAg and the emergence of anti-HBsAg antibodies indicate the durable immune control of HBV infection or the functional cure (Nguyen et al., 2020). Hence, knowledge on the mechanism of HBV envelope protein metabolism and sSVP morphogenesis is important for the rational discovery and development of antiviral drugs to achieve this therapeutic goal. Besides, sSVPs are used as HBV vaccines for decades.
All available data suggest that sSVP hijacks the classic secretory pathway of the hepatocyte including the key organelles encompassing the ER, the ERGIC, the Golgi apparatus and the plasma membrane, along with interconnected vesicular traffic devices and routes (Chuang et al., 2022; Prange, 2012; Seitz et al., 2020). The HBV S envelope protein alone is sufficient for the production and secretion of sSVPs, defined lipoprotein complexes that are formed by self-assembly and intraluminal budding of about 96 transmembrane S molecules. Essentially, two models are discussed concerning the mode and the location of the self-assembly process. Very early works provided biochemical and cell biological evidence that S is cotranslationally translocated into the ER membrane (Eble et al., 1986). One model proposes that transmembrane S monomers rapidly form disulfide-linked dimers catalyzed by the ER chaperone protein disulfide-isomerase (PDI). Transmembrane dimers are next transported to the ERGIC where the absence of PDI and a different environment allow intraluminal SVP budding (Huovila et al., 1992). Another model is derived from electron microscopic studies and predicts that S self-assemble initially in a filamentous form within the ER. These filaments are folded, bridged into crystal-like structures, and then transported to the ERGIC where the filaments are unpacked before being converted into spherical particles for secretion (Patient et al., 2009). As outlined above, the HBV envelope is exported out of the ER in a COPII-dependent manner, irrespective whether it ends up in virions or sSVPs. To coopt COPII, S specifically interacts with the Sec24A isoform with aid of its cytosolically disposed di-arginine-78/79 motif (Zeyen et al., 2020). The direct S-Sec24A interaction suggested that Sec24A recognizes transmembrane S chains for COPII-guided transport to the ERGIC and, in turn, that intraluminal SVP budding should occur beyond the ER. Similar conclusions were drawn from a study that uses pharmacological disruption of vesicle trafficking between the ER and Golgi complex in sSVP producing cells (Yang et al., 2021). Moreover, this study identified a novel, amphipathic α-helix spanning aa 156–169 of S that plays essential roles in sSVP production. Possibly, this helix may embed in the lumen leaflet of ERGIC membranes, which may alter the membrane curvature and/or lipid fluidity and consequentially facilitate sSVP budding (Yang et al., 2021). Following budding, the sSVPs are transported through the Golgi stacks, where the N- glycan, linked to Asn-146, is processed from the high-mannose type to the complex type and subsequently secreted from hepatocytes via exocytosis. Notably, the cellular ERGIC-53 lectin plays no role in sSVP trafficking (Zeyen et al., 2020), and the Golgi complex is important for sSVP secretion but not production.
Since a functional cure of CHB requires eliminating the immune suppressing spheres, sSVPs have gained great attention as antiviral targets. Candidate compounds include nucleic acid polymers (NAPs), cell-permeable phosphorothioate oligonucleotides that can inhibit sSVP assembly/release in vitro and in vivo. Since NAPs can clear sSVPs from the blood without any discernible direct interaction with HBV components, they are thought to target host proteins selectively involved in viral envelope trafficking (Boulon et al., 2020). Recently, a new class of NAPs called S-antigen transport-inhibiting oligonucleotide polymers (STOPS) were developed that are able to inhibit HBV infection and HBsAg production in vitro. Again, they do not bind HBV molecules, but rather targets host factors, including the BiP chaperone, RPLP1, and RPLP2 (60S acidic ribosomal protein P1 and P2), that are required for the proper translation and folding of S/sSVPs (Kao et al., 2022).
In view of the little number of reports, filamentous fSVPs are a poor cousin of spheres. Again, the L protein is the key determining the formation of filaments that are frequently observed in vivo with length ranging from 200 nm to 1.2 μm. Like viral particles, filaments are characterized by a significantly higher content of L as compared to spheres. They are estimated to contain L, M, and S in the ratio of 1:1:4, respectively (Heermann et al., 1984) that assemble as a mixed population of homo- and heterodimers into tubular particles with spike-like features projecting from the membrane (Short et al., 2009). To study filament dynamics and trafficking, mutant HBV replicon constructs had been used that were unable to produce the core protein. Cell imaging studies of these cells showed that L, concomitant with filamentous particles, accumulated in a crescent-shaped, probably ER-related structure in perinuclear regions (Jiang et al., 2015). Very surprisingly, Jiang et al. (2015) showed that filaments share the viral export route as they also require ESCRT functions – in particular ESCRT-III and Vps4 – for cell exit. Since α-taxilin, an interaction partner of the ESCRT-I-specific Tsg101 protein, was likewise essential for the release of filaments, the authors concluded that L may use this subunit to enter the ESCRT/MVB machinery (Hoffmann et al., 2013). However, filament export via the MVB portal again raises the question how and when the N-linked carbohydrates of L are processed within the Golgi complex. Curiously, the length of filaments is determined by a preS1-specific subregion within aa 25–39. A corresponding envelope deletion was isolated from a patient suffering from chronic HBV infection. Beside affecting the size of filaments, the deletion led to the appearance of semi-enveloped virions, that is, incompletely covered nucleocapsids (Jiang et al., 2019). Such a phenomenon has been unprecedented so far, but the partial core exposure may eventually explain how high titers of anti-core antibodies are produced in virtually all patients who have been exposed to HBV.
Notably, also nonenveloped or naked capsids/NCs can be secreted in cell-culture models, but had been considered to be absent in vivo (Bardens et al., 2011; Sun & Nassal, 2006; Watanabe et al., 2007; Wittkop et al., 2010). A recent work could perhaps provide an explanation for his contradiction by demonstrating that such capsids may be complexed with anti-capsid antibodies and thereby undermine detection in patients (Bai et al., 2018). Conversely, however, an earlier study had shown that no naked HBV capsids were detectable in immunosuppressed patients, that is, even in the absence of anti-HBc antibody (Possehl et al., 1992). Moreover, in animal models, like immunocompromised HBV-transgenic mice or HBV-infected humanized chimeric mice, only very low levels of naked capsids were detectable (Hong et al., 2021). Although the physiological relevance of naked capsids must be confirmed beyond doubt, they may carry HBV-specific DNA and RNA species (Deng et al., 2022; Liu et al., 2019) and have been shown to be competent for clathrin-mediated endocytosis (Cooper & Shaul, 2006). Accordingly, their uptake may enhance transmission of HBV genomes. Recently, it was found that nackednaviruses, which were separated from hepadnaviruses over 400 million years ago, do not encode any envelope proteins and secrete viral particles in forms that are comparable to the naked capsids in HBV (Lauber et al., 2017). This reinforces the idea that the release of unenveloped nucleocapsids is a natural process of viral egress, not the result of passive cell lysis. In support, cell-culture based analyses revealed that the non-lytic release of naked HBV capsids is supported by the cellular Alix protein, a multifunctional protein with key roles in membrane structure and biology. Ectopic overexpression of Alix or even its N-terminal, boomerang-shaped Bro1 domain, which have membrane-modulating activities, enhanced capsid egress (Bardens et al., 2011; Watanabe et al., 2007). Besides, the ESCRT-0 subunit HRS is a promoter of naked capsid egress while the classic ESCRT machinery is not involved (Chou et al., 2015).
The recent discovery of genome-free, empty viral particles demonstrates that HBV has even more surprises in store. Such incomplete HBV particles contain both the envelope and capsid but no genome and are also secreted from HBV-replicating hepatocytes, both in vivo and in vitro (Hu & Liu, 2017). Similar to empty envelopes, incomplete particles are released in excessive amount as compared to virions and are thought to perform immune modulatory functions. Since these particles contain all HBV structural proteins, but are non-infectious, they are discussed as a next generation vaccine candidate. As summarized above, the discovery of genome-free particles led to the questioning of the maturation signal hypothesis (Ning et al., 2011). The analysis of capsid and envelope protein requirements unexpectedly revealed that the L protein was not required for empty virion secretion (Ning et al., 2018) and in such suggested an ESCRT- and MVB-independent export pathway. Why HBV releases such an excessive amount of non-infectious particle types remains a fascinating but challenging question still today. But confronting the host with several distinct particle types in huge amounts might also be one of the reasons for HBV's immense success as a human pathogen.
ACKNOWLEDGMENTS
I am grateful to past and present lab members for their contributions. Work cited from the author´s lab has been or is supported by grants from the Deutsche Forschungsgemeinschaft (SFB 490-D1, PR 305/1-3, PR 305/3-1, PR 305/3-2, PR 305/3-3).
Open access funding enabled and organized by Projekt DEAL.
CONFLICT OF INTEREST
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.