A machine learning-based workflow for predicting transplant outcomes in patients with sickle cell disease
Haiou Li, Vandana Sachdev, Colin O. Wu and Swee Lay Thein contributed equally.
Summary
Allogeneic haematopoietic cell transplantation (HCT) with HLA-matched sibling donor remains the most established curative therapeutic option for patients with sickle cell disease (SCD). However, it is not without risks, highlighting the need for a risk stratification system. Utilizing a machine learning (ML) approach that combines clinical and imaging variables, we identified red cell distribution width and renal organ damage as important risk factors for patients undergoing HCT. This ML-based algorithm, similar to an approach previously reported for predicting mortality in patients with SCD, should be applicable to risk factor discovery in similar studies.
INTRODUCTION
Sickle cell disease (SCD), caused by a single-point mutation in the β-haemoglobin gene, is probably the most common severe inherited disease in the world.1 The clinical manifestations are multisystemic with progressive organ damage and premature mortality. Current drug therapies suffer from poor patient adherence and are not effective in all patients. Recently, the US Food and Drug Administration (FDA) approval of two genetic therapies—Casgevy and Lyfgenia—has increased therapeutic options,2 but allogeneic haematopoietic cell transplantation (HCT) with HLA-matched sibling donor remains the more established and relatively more accessible option for patients. Nonetheless, these curative interventions are high-risk procedures involving potentially significant toxicities and organ complications.2-5 In 2021, the American Society of Haematology published a guideline with eight recommendations on HCT for SCD, but it was acknowledged that the evidence for all recommendations is of low certainty due to a lack of randomized clinical trials.6 A lack of universal end-points in HCT trials was also noted, highlighting an immediate need for predictive biomarkers of mortality associated with HCT. In prior work, we developed a phenotypic risk score to predict mortality in SCD,4 and we demonstrated that our machine learning (ML)-based approach had superior performance compared with competing models. However, we recognized that SCD patients who underwent HCT were censored and not included in the analysis. Here, we applied a similar ML approach to develop a risk stratification workflow to predict both mortality and transplant outcomes specifically in adults with SCD undergoing HCT.
METHODS
Study design and participants
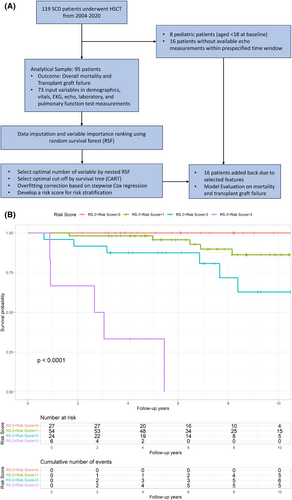
The study included SCD patients who underwent HCT (full HLA-matched siblings and haploidentical, Table S1) at the NIH Clinical Centre, between September 2004 and February 2020 (Clinicaltrials.gov: NCT#00061568, NCT#00977691, NCT#02105766 and NCT#03077542) (Figure 1). All transplant protocols were approved by the National Heart, Lung, and Blood Institutional Review Board, and all subjects signed informed consent prior to participation. Baseline echocardiogram (echo) and laboratory measurements were defined as those performed within 30 days of the consent date, and the final follow-up for all patients was in March 2023. Of 119 patients, 17 were originally excluded, 16 due to lack of echo data, and one due to lack of laboratory data. Seven patients aged <18 years were excluded. Initially, 97 covariates (demographics, echo, electrocardiogram (EKG), vitals, laboratory, and pulmonary function tests (PFTs)) were considered as candidate risk factors for mortality and transplant failure (Table S2); however, those with >30% missing values were excluded resulting in 73 variables that were included in the analysis (Figure 1).
Study outcomes
The primary outcome was all-cause mortality ascertained by the date between the consent date and the date of the last follow-up, proxy interview, medical records, National Death Index (NDI), and Social Security Death Index (SSDI) search. Transplant failure was defined as graft failure by the return of acute sickle-related complications, including acute painful episodes, acute chest syndrome, or anaemia, with or without evidence of haemolysis. A secondary analysis was conducted based on the composite transplant outcome, of which event was identified by transplant failure or death (whichever occurred first).
Statistical analysis
We used a two-step statistical ML approach to select the top influential covariates for mortality (see previous study7), which combined the process of random forest (RF), nested RF, and stepwise Cox proportional hazard model. The feature selection was based on 95 patients and the ML-model was evaluated on all patients with available final selected risk factors (N = 111). Details of the ML methods and statistical analyses are available in the Appendix S1.
RESULTS
Characteristics of the study cohort
A total of 111 SCD (106 HbSS and HbSβ0 thal) patients underwent HCT, 74 (67%) with a fully HLA-matched related donor and 37 (33%) with a haploidentical donor (Table S1). The median age of participants was 31.8 (24.7–37.8) years, 38% were female, other demographic details are shown in Table S1. Over a median follow-up of 6.9 years (maximum follow-up of 19.0 years), 17 deaths were observed (median age [IQR] 40 [27–47] years) and the 4-year mortality was estimated at 7.2% (95% confidence interval [CI] 2.3%–12.0%) (Figure S1), compared with 14% in the general SCD population.7 For the HCT procedure, 27 graft failures (24.3%) were observed during a median time of 5.24 years after the transplant.
Machine learning for ranking variable importance
Using the model building cohort of 95 patients with 73 covariates (Figure 1), variable importance (VIMP) was calculated by the random survival forests (RSF) method for mortality prediction (Figure S2A). Based on the VIMP ranking, a nested RSF was applied which suggested that the optimal number of variables to reach the maximal model performance was eight achieved by eight covariates. These top eight predictors were blood urea nitrogen (BUN), creatinine, left ventricular posterior wall (LVPW) diastolic wall thickness, lactate dehydrogenase (LDH), red blood cell distribution width (RDW), estimated right ventricular systolic pressure (RVSP), absolute reticulocyte count and direct bilirubin (Figure S2B).
Cut-off point and risk score development based on stepwise Cox regression
The top eight selected risk variables were entered into a multivariable model (model 1) with a resulting a C-statistic of 0.831. In order to develop an easy-to-apply risk classification, the optimal cut-off points were chosen for each of eight risk factors based on the survival classification and regression tree8 (CART) (Table S3 and Figure S3). A stepwise Cox proportional hazard regression was further applied to correct for potential overfitting, which identified four independent predictors—BUN, LDH, RDW, and absolute reticulocyte count—that were associated with all-cause mortality. Model 2, based on these four categorial risk factors, performed better with a higher C-statistic of 0.840 compared with Model 1 (Table S4). In Model 3, we assigned equal weight to each of the 4 predictors and compiled a risk score as a sum of all the binary factors in the cohort (Table S4). Performance of the simplified risk score using the four categorized factors was similar compared with the model using eight factors with C-statistic = 0.842 (Figure S4A). We applied the HCT-Comorbidity Index (HCT-CI) risk stratification tool to our cohort and found that our new phenotypic risk score had better discrimination and predictive capacity (C-statistic, 0.842 vs. 0.679, Figure S4B).9, 10
Since the top four predictors in the phenotypic risk score did not include echo variables, we reinstated the 16 transplant subjects that were initially excluded (due to lack of echo data) and repeated the analyses. The C-statistic of 0.815 using Model 3 on the cohort of 111 illustrates the robustness of our developed risk scoring system and 4-year mortality estimates were 0%, 3.2%, 7.9%, and 66.7% based on risk scores of 0, 1, 2, and 3, respectively (Figure 1B). The significance remained when controlling for the transplant donor type (C-statistic 0.871, Figure S5).
For the secondary analysis, amongst the 111 subjects in the full cohort, a higher mortality was observed in the 27 who failed HCT compared with those who engrafted (40.7% vs. 7.1%). Besides the mortality stratification, the risk scores of 0, 1, 2, and 3 could also identify the patients' risk of transplant failure, not taking into account the donor type (C-statistic 0.649) with 4-year disease progression estimates of 9.1%, 22%, 32.8%, and 74.1% (Figure S6; Table S5). Accounting for the type of transplant donor improved C-statistic to 0.769 (Figure S7; Table S5).
DISCUSSION
Considering the significant risks associated with HCT in SCD patients, the development of a risk stratification system to identify patients at the highest risk of early mortality and significant morbidity after HCT, is a priority. This system could serve as both an assessment and decision-making tool to aid in determining eligibility for curative therapies. Furthermore, developing an algorithm that aligns the risks of graft failure and survival after HCT with the SCD disease severity will facilitate monitoring of patient disease progression for timely HCT intervention. Based on a single-centre cohort of 111 SCD patients, our ML-based workflow selected a set of baseline biomarkers, alongside the transplant donor type, that were associated with both SCD mortality and transplant outcomes. Four critical and readily accessible clinical variables—BUN, LDH, RDW, and absolute reticulocyte count—were identified and the risk models utilizing these risk factors showed enhanced performance in predicting the risk of mortality and graft failure.
The variable identified as having the greatest importance by RSF was BUN, a global biomarker of renal complications in SCD, and it was found to be more predictive in our study than other biomarkers of renal function, such as serum creatinine, eGFR, and urine albumin/creatinine ratio.7 Lactate dehydrogenase has previously been shown to be associated with the severity of vaso-occlusive episodes and muti-organ failure, as well as survival in SCD.11, 12 Our current findings underscore the importance of routine blood laboratory parameters as prognostic indicators, particularly absolute reticulocyte count and RDW, which have been associated with acute SCD pain episodes13 and vascular pathology14 in transplant outcomes.
The proposed risk prediction workflow (Figure 1A and Figure S8), which integrates ML techniques for biomarker selection and model building, outperformed the established HCT-CI index in predicting outcomes for our SCD transplant cohort. A main limitation of the current study is the lack of a validation cohort for our risk prediction algorithm, which was derived from a limited, single-centre cohort. Interval validation based on bootstrap and sample splitting approach was applied to evaluate the performance of the novel risk score and proposed workflow. However, given the heterogeneity in SCD severity and patient phenotypes amongst different centres, we recommend implementing the ML workflow, rather than the specific biomarkers identified, for tailored centre-specific risk identification and transplant recommendation (Figure S8). This workflow is both versatile and adaptable for risk stratification and the selection of SCD transplant candidates across various centres.
AUTHOR CONTRIBUTIONS
Vandana Sachdev, Haiou Li, Colin O. Wu, and Swee Lay Thein designed the study. Haiou Li and Vandana Sachdev contributed equally as the first authors. Colin O. Wu and Swee Lay Thein contributed equally as senior authors. My-Le Nguyen, Matt Hsieh, Courtney Fitzhugh, Emily Limerick, Nancy Asomaning, and Anna Conrey contributed to data acquisition, curation, and analysis. Haiou Li and Vandana Sachdev verified the underlying data. Haiou Li, Vandana Sachdev, Xin Tian, and Colin O. Wu contributed to the statistical analysis. Haiou Li, Vandana Sachdev, Xin Tian, Colin O. Wu, and Swee Lay Thein wrote the first draft of the manuscript. All authors critically reviewed the draft and approved the final version for publication.
FUNDING INFORMATION
This research was supported by the Intramural Research Programme of the National Heart, Lung, and Blood Institute, NIH, Department of Health and Human Services (DHHS).
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests.
ETHICS STATEMENT
All protocols were approved by the NHLBI Institutional Review Board. Written informed consent was obtained from all individuals enrolled in the study.
CLINICAL TRIAL REGISTRATION (INCLUDING TRIAL NUMBER)
Clinicaltrials.gov: NCT#00061568, NCT#00977691, NCT#02105766 and NCT#03077542.
Open Research
DATA AVAILABILITY STATEMENT
The datasets generated and/or analysed in the current study are available from the corresponding author upon reasonable request.




