Single gene mutations and prognosis in limited-stage follicular lymphoma treated with radiation therapy
Summary
Radiotherapy is routinely used for management of limited-stage follicular lymphoma (FL), yet half of patients ultimately relapse. We hypothesized that the presence of specific gene mutations may predict outcomes. We performed targeted sequencing of a 69-gene panel in 117 limited-stage FL patients treated with radiotherapy and identified recurrently mutated genes. CREBBP was most frequently mutated, and mutated CREBBP was associated with inferior progression-free survival, though not after false discovery rate adjustment. This association failed to validate in an independent cohort. We conclude that recurrent gene mutations do not predict outcomes in this setting. Alternative biomarkers may offer better prognostic insight.
INTRODUCTION
Limited-stage follicular lymphoma (FL), defined as stage I or II disease, is generally treated with radiation therapy (RT) to involved nodes with excellent overall response.1-3 However, nearly half of all limited-stage FL patients will ultimately experience relapse.4, 5 There are differences between international guidelines with respect to treatment options, with some suggesting systemic treatment with chemoimmunotherapy is a reasonable alternative, based on studies demonstrating improved progression-free survival (PFS), but not overall survival (OS).2, 3, 6, 7 In other lymphoma subtypes, there is evidence that gene mutations impact survival and may predict treatment response.8 This prompted the question of whether we could guide treatment in limited-stage FL by genetic mutations, specifically, whether we could identify a subset of patients through genetic means who have poor prognosis with radiotherapy and may benefit from systemic treatment.
Current prognostication for FL is done by the Follicular Lymphoma International Prognostic Index (FLIPI), which is based on clinical variables. Most prior work with genetic prognostication in FL has focused on patients with advanced disease undergoing systemic therapy. Pastore et al. developed a clinicogenetic risk model, the m7-FLIPI, which incorporated mutation status of seven genes, FLIPI score and ECOG performance status in a cohort of advanced FL patients receiving chemoimmunotherapy.9 They found that risk stratification using m7-FLIPI improved prognostication compared to FLIPI alone.
However, there is evidence to suggest genetic differences in limited versus advanced-stage FL, for example, in frequency of KMT2D and BCL2 mutations and gene expression profiles.10-13 Taken together, we expected there may be differences in which mutations drive disease and prognosis in limited versus advanced FL.
We, therefore, hypothesized that genetic characterization of pre-treatment biopsies could identify those limited-stage FL patients who are at high risk of relapse following radiation therapy.
PATIENTS AND METHODS
Pre-treatment formalin-fixed and paraffin-embedded tissue block samples were obtained from adult patients with limited-stage FL, grades 1–3A, treated with curative-intent RT diagnosed between 1995 and 2018, from six international sites in Canada, Europe and Australia. Hybridization-based targeted sequencing of a 69 gene panel was performed, which included genes known to be recurrently mutated in FL (Table S1). Samples with >50X mean on target coverage were retained in the analysis. Variant calling was performed using Mutect2.14, 15 Germline polymorphisms were filtered out by cross-referencing with dbSNP and gnomAD databases.16, 17 Only coding, non-silent mutations detected at variant allele frequency >0.1 were considered. Patient demographics and clinical data were examined retrospectively. Genes mutated at a proportion of 10% or more of the cohort were tested for association with PFS and OS.
Based on our initial findings, a separate cohort of patients consisting of limited-stage FL patients treated with radiotherapy alone as part of the TROG 99.03 randomized clinical trial was subsequently used to explore the relationship between CREBBP mutation and clinical outcomes.7 TROG 99.03 was a randomized, international, multicentre phase III trial comparing involved-field radiation alone to radiotherapy followed by chemoimmunotherapy in stages I–II FL. A distinct targeted sequencing panel was applied to FFPE tumour samples from this independent cohort but CREBBP variants were called using the same methods described above.
RESULTS
Overall, 117 pre-treatment samples from distinct patients were accrued. The median age was 58 years (range: 26–86), and 68 (58%) were females (Table 1). There were 99 (85%) patients with stage I disease, and 18 (15%) with stage II. Grade was 1 or 2 in 87% of patients. Mean maximum tumour diameter was 3.33 cm (range: 0.5–8 cm). Of 110 patients with FLIPI scores available, 100 patients had a low FLIPI score (91%), nine (8%) had an intermediate score and one (1%) patient had a high score. Patients were staged with cross-sectional imaging, including a PET scan in 41 (35%) of cases, and bone marrow assessment was completed in 105 (90%) of patients.
| Total n = 117 | |
|---|---|
| Age (median, range) | 58 (28–86) |
| Sex | |
| Female | 68 (58%) |
| Male | 49 (42%) |
| Stage | |
| I | 99 (85%) |
| II | 18 (15%) |
| Grade | |
| 1 or 2 | 101 (87%) |
| 3a | 15 (13%) |
| Maximum tumour diameter (mean, SD) | 3.3 cm (1.48) |
| FLIPI | |
| Low | 100 (85%) |
| Intermediate | 9 (8%) |
| High | 1 (1%) |
| Unknown | 7 (6%) |
| PET staging | |
| Yes | 41 (35%) |
| No | 73 (62%) |
| Unknown | 3 (2%) |
| Bone marrow assessment | |
| Yes | 105 (90%) |
| No | 11 (9%) |
| Unknown | 1 (1%) |
- Abbreviations: FLIPI, follicular lymphoma international prognostic index; PET positron emission tomography.
The most commonly mutated gene in the cohort was CREBBP (n = 61; 52.1%), followed by KMT2D (n = 45; 38.5%), TNFRSF14 (n = 40; 34.2%), STAT6 (n = 25; 21.4%) and BCL2 (n = 20; 17.1%; Figure S1). BCL2 mutations were more common in patients with stage II (n = 8; 44%) compared to stage I disease (n = 12; 12%; p-value = 0.003; after false discovery rate (FDR) adjustment, p-value = 0.059; Figure S2). The median number of mutated genes per patient was 4 (range: 0–12, mean 4.0), with no significant difference between stages I and II (p-value = 0.14). The median follow-up of living patients was 6.4 years (range: 0.63–21.7 years) and there were 50 progression events in our cohort.
CREBBP mutations were associated with shorter PFS (log-rank p-value 0.012), with 5-year PFS estimates of 66.8% versus 82.0% (Figure 1A). CREBBP mutations were not significantly associated with age, stage, tumour bulk or PET staging. Using Cox univariate analysis for PFS, the hazard ratio (HR) for CREBBP mutations was 2.1 (95% confidence interval (CI) 1.2–3.7; p-value = 0.0145) and in multivariable analysis adjusting for age, stage and sex the HR was 2.56 (95% CI 1.37–4.80; p-value = 0.0032). However, statistical significance was not met after adjusting for the FDR (p-value = 0.13). CREBBP mutations were not significantly associated with OS (log-rank p-value = 0.66; Figure 1B), nor transformation (log-rank p-value = 0.54), but the numbers of survival (n = 20) and transformation events (n = 11) were small. No other genes from the panel were associated with patient outcomes.
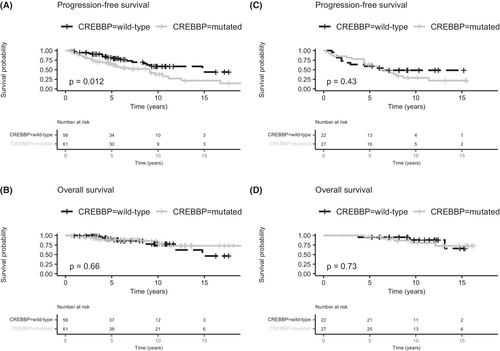
The extension cohort consisted of 49 patients with CREBBP mutation data available from the radiotherapy-alone arm of the TROG 99.03 clinical trial.7 There was no significant difference in either PFS (p-value = 0.43) or OS (p-value = 0.73) by CREBBP mutation status (Figure 1C,D).
A subgroup analysis of patients with missense mutations in the histone acetyltransferase (HAT) domain of CREBBP was performed, as this domain harbours the majority of CREBBP mutations in FL.19 Additionally, missense mutations in particular within the HAT domain may have distinct downstream effects and are more prevalent in FL compared to other mutation types.18 Sixty-two percent (38 out of 61) of samples with CREBBP variants had HAT domain missense mutations and these were associated with inferior PFS compared to other types of CREBBP mutations or wild-type status (p-value = 0.034; Figure S3). This association was not statistically significant in the extension cohort (p-value = 0.37; Figure S4).
An exploratory analysis examining patients with stage I disease only for associations with PFS was performed. In Cox univariable analysis, the HR for CREBBP mutations was 2.3 (95% CI 1.1–4.6; p-value = 0.019), and the HR for BCL2 was 2.6 (95% CI 1.1–6.1; p-value = 0.025), however, neither remained significantly associated with PFS after FDR correction (p-value = 0.11 for both).
DISCUSSION
Though we initially found an association between CREBBP mutations and shorter PFS in our cohort, this did not persist after FDR correction and was not validated in an external population. More generally, no associations existed between any other gene on our 69-gene panel and outcomes.
This is consistent with a prior study by Kalmbach et al., who performed whole-exome sequencing of 147 stages I–II FL cases.20 The most frequently mutated genes in their limited-stage cohort were similar to our study with comparable CREBBP and KMT2D mutation frequencies. However, CREBBP mutations were more prevalent than KMT2D in our cohort. Additionally, they reported ARID1A mutations being less common in limited-stage (6%) versus advanced-stage FL (29%). In our cohort, ARID1A mutation frequency was similarly low (9%). Outcome correlation was a secondary objective of Kalmbach et al., and they analysed a smaller panel of six genes (five are included in our panel) for association with PFS.20 Similar to our results, they found no association between single gene mutations, including CREBBP, and PFS.
The m7-FLIPI clinicogenetic risk model incorporated seven genes (all of which were included in our panel) and FLIPI score.9 In univariate analysis, EP300 and FOXO1 mutations were associated with higher risk and EZH2 mutations with lower risk of failure-free survival. However, similar to our results, after correction for multiple testing, no genes were associated with failure-free survival in the 74 genes examined.
Russler-Germain et al. identified a seven-gene signature, which included CREBBP, where the presence of ≥2 mutations was associated with inferior PFS in FL. However, only 21% of included patients had limited-stage, and no patients were treated with radiotherapy alone.21 Gao et al. found that HIST1H1D mutations were associated with progression; however, only 22% had limited-stage, and less than 5% of the population were treated with radiotherapy alone.22 It is unclear whether these results apply to limited-stage patients treated with radiotherapy.
Los-de Vries et al. described three clusters using genomic and immune microenvironmental data in stage I FL, with two of these clusters being unique to stage I compared to stages III–IV disease.13 There is greater heterogeneity in clinical presentation and treatment choice in stage II versus I FL, and it is unclear whether there are biological differences. We did find a higher frequency of BCL2 mutations in stage II compared with stage I patients, though in an exploratory analysis including only stage I patients, there were no associations between any genes on the panel and PFS or OS after FDR adjustment.
Limitations of our study include limited sample size, given the large number of genes tested for association. Additionally, considering the excellent prognosis of limited-stage FL, the follow-up was relatively short. PET staging, which can result in upstaging of disease, was not uniformly performed in all patients. BCL2 translocation status was not routinely collected and unavailable in most patients, and thus was not reported.
In conclusion, it appears that single gene mutations, or at the very least, the panel examined in our study, are likely not the main driver of prognosis in limited-stage FL. Future studies examining prognosis in limited-stage FL may better focus on other factors such as tumour microenvironment, gene expression profiles or other genetic mutations not included in our panel.
ACKNOWLEDGEMENTS
S.A.H., J.T., V.S., M.K.G., R.K. performed the research. R.K. designed the research study. J.T., L.D., K.L., T.L., M.B., F.A.D., M.L., T.B., D.L., N.J., M.C., M.H., J.K., R.T.L., R.T., D.H., M.M., M.K.G. contributed essential materials. S.A.H., J.T., V.S., K.L., M.K.G., R.K. analysed the data. S.A.H., R.K. wrote the paper.
FUNDING INFORMATION
This work was supported by the Leukemia and Lymphoma Society of Canada. S.A.H. was supported by the Hold ‘em for Life Oncology Fellowship.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest associated with this work.
ETHICS APPROVAL STATEMENT
Ethics approval has been obtained from the University of Health Research Ethics Board, CAPCR ID: 17-5089.
Open Research
DATA AVAILABILITY STATEMENT
The sequencing data have been deposited at the European Genome-phenome Archive (https://ega-archive.org/, accession number EGAS0000000435).




