A nonsense variant in the Hexokinase 1 gene (HK1) causing severe non-spherocytic haemolytic anaemia: genetic analysis exemplifies ambiguity due to multiple Isoforms
Hexokinase (HK) catalyses the first, rate-limiting step of the glycolytic pathway, converting glucose to glucose-6-phosphate. Mammalian HK has four main isozymes: HK1, HK2, HK3 and HK4, encoded by different genes. HK1 is ubiquitously expressed, but with predominant expression in the brain, erythrocytes, testis and other tissues where glycolysis serves as the major source of ATP production. Erythrocytes express two major isoforms formed by the alternative splicing of the HK1 gene: HK-R (NM_033496, NP_277031) and HK-1 (NM_000188, NP_000179) (Murakami et al, 2002). HK deficiency (OMIM # 235700) is a rare erythroenzymopathy with an autosomal recessive mode of inheritance, and is associated with mild to severe non-spherocytic haemolytic anaemia. Fetal death has been reported in one case (Kanno et al, 2002). Although approximately 30 cases of HK deficiency have been reported, molecular findings have been described in only 11 patients (Khazal et al, 2016; Koralkova et al, 2016). Definitive diagnosis requires biochemical testing of enzyme levels and/or genetic testing, which are often difficult and not readily available.
A nine-month-old boy from central India was referred for severe anaemia since birth (requiring 14 blood transfusions, starting with an exchange transfusion at day 4 of life for neonatal jaundice), recurring jaundice and delayed milestones. The infant had increasing abdominal distension for the past three months. He was born of consanguineous marriage (maternal cousins); both sets of grandparents were also first cousins. Four male siblings of his mother had died within 10–15 days of the birth of undocumented causes. There was no blood group or Rh incompatibility between mother and child, and there was no family history of splenectomy or cholecystectomy. On examination, the liver and spleen were palpable 7 and 12 cm below the respective costal margins.
Complete blood counts revealed haemoglobin 47 g/l (120–180), red blood cell (RBC) count 1·47 × 1012/l (3·8–5·5), mean corpuscular volume 92·3 fl (80–96), mean corpuscular haemoglobin 31·8 pg (27–32), mean corpuscular haemoglobin concentration 345 g/l (300–360), red cell distribution width (coefficient of variation) 16·6% (11·6–14), total corrected leucocyte count 22·6 × 109/l, platelet count 40 × 109/l and reticulocytosis (12·8%). RBCs were predominantly normocytic normochromic with mild anisocytosis, polychromasia, occasional spherocytes, basophilic stippling and many Cabot rings (Fig 1A). A peripheral smear had a leucoerythroblastic picture showing 5% myelocytes and 21 nucleated RBCs/100 leucocytes with mild dyserythropoiesis (Fig 1B, C).
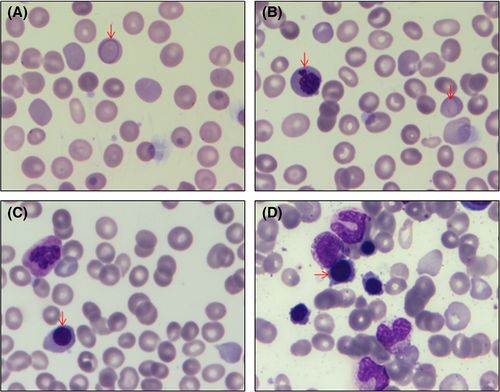
The clinical differential diagnoses included inherited haemolytic anaemia and myelofibrosis. Other causes of haemolytic anaemias were excluded as described before (Jamwal et al, 2017). Briefly, haemoglobin high-performance liquid chromatography, methaemoglobin reduction test, pyruvate kinase enzyme activity assay, flow cytometric eosin-5′-maleimide dye binding test, Heinz body preparation, direct antiglobulin test, and isopropanol and heat stability test were all normal/negative. Incubated osmotic fragility test showed increased resistance to osmotic lysis, probably due to reticulocytosis. Bone marrow was normocellular with erythroid hyperplasia and no storage cells or fibrosis (Fig 1D).
To further determine the diagnosis, DNA from the patient, his parents and asymptomatic sibling was subjected to targeted resequencing using TruSight One Sequencing Panel (Illumina Inc., San Diego, CA, USA) after obtaining the necessary approval and consents. Preparation of DNA libraries and data analysis used MiSeq Reporter and VariantStudio, as described (Jamwal et al, 2018), but no potentially pathogenic variant was found. For whole-exome sequencing (WES), DNA libraries were prepared using the Nextera Rapid Capture Enrichment guide (Illumina) by enzymatic fragmentation as per the manufacturer's instructions (tagmentation) followed by purification to ensure uniform and appropriate fragment size. The purified fragments were ligated to adapters specific for the Illumina HiSeq platform. For target enrichment of the exome, the library was hybridized to biotinylated probes. The pool was enriched for the desired region by adding streptavidin beads. Unbound DNA was washed away and enriched fragments were eluted and further amplified by polymerase chain reaction (PCR). Prepared libraries were sequenced on the HiSeq 2500, and the resulting FASTQ data files subjected to quality assurance/quality control. Sequence reads were mapped to HG19 sequence and variants were identified by a bioinformatics pipeline developed at National Institute of Biomedical Genomics, Kalyani (India Project Team of the International Cancer Genome Consortium, 2013).
Data analysis revealed a homozygous nonsense variant NM_033496.2:c.34C>T, p.Arg12Ter, in exon 1 of HK1 (rs756166032) (Fig 2A) that had a minor allele frequency of T = 0·00002/3 in the Exome Aggregation Consortium database (http://exac.broadinstitute.org/). Both parents and his asymptomatic sister were heterozygous for this substitution. This was tested using PCR-restriction fragment length polymorphism as the variant results in loss of a TaqI restriction endonuclease site (forward primer ACTAAGGACTCCCTGTGGGT and reverse primer TCGCCGCTCATATTCCCTTC; product size 623), confirmed an autosomal recessive mode of inheritance (Fig 2B).

Although the case was tested via targeted resequencing before WES, the variant was missed during data analysis. Re-evaluation of the data revealed that there was different annotation used for the variant in the VariantStudio (Illumina), which was probably based on the different isoform of the HK1 gene. HK1 is one of the ubiquitously expressed isozymes among mammalian hexokinases, which are all encoded by different genes (HK1, HK2, HK3 and HK4 on chromosomes 10, 2, 5 and 7 respectively). Furthermore, HK1 has multiple isoforms i.e., HK-1 (ubiquitous, NM_000188), HK-R (erythrocytes, NM_033496), HK-TA (testis-A, NM_033497), HK-TB (testis-B, NM_033498), HK-TD (testis-D, NM_033500), etc. which are formed by the alternative splicing of the exons. HK-1 and HK-R isoforms differ only in exon 1 and share the remaining 17 exons. Isoforms HK-TA, HK-TB, HK-TC and HK-TD have different 5′ UTR exons and share the rest 17 exons with all other isoforms of HK1 (Figure S1).
RBCs express two major isoforms HK-R and HK-1 (Murakami et al, 2002). In our case, the HK-TA (NM_033497) isoform was set as default in VariantStudio, which interpreted the variant as intronic (NM_033497.2:c.75+20308C>T), and was thus overlooked during data analysis (Fig 2C). This case illustrates that variants can be annotated based on different isoforms and that automated data analysis should only take into account the isoform expressed in the implicated/suspected disorder(s). As targeted resequencing is more often based on automated systems in clinical practice, a near-error situation may arise, as exemplified here. Unfortunately, the child succumbed at one-year of age. The family has been counselled for prenatal screening in future pregnancies.
HK deficiency is a very rare red cell enzymopathy and, to the best of our knowledge, no case from India has been reported at the phenotypic or genetic level. The conventional biochemical enzyme assay to estimate the HK activity in RBCs is often confounded by recent blood transfusions and reticulocytosis. Moreover, as compared to other glycolytic enzymes, which gradually decline during the life span of erythrocytes, HK declines in a biphasic manner. HK-R declines rapidly, has maximum activity in young reticulocytes and is absent in older erythrocytes, however, HK1 declines gradually over the life span of erythrocyte (de Vooght et al, 2009). Therefore, the timing of the assay after sample collection is also important, adding to the difficulty of diagnosing the disorder. A gene-by-gene approach is also expensive, time-consuming and cumbersome because there is no pathognomic RBC morphological finding for HK deficiency. Hence, molecular diagnosis using next-generation sequencing with careful data analysis can provide an accurate diagnosis in such cases and prenatal diagnosis can be offered due to the severity of the clinical phenotype.
Author contributions
M.J., A.A., P.S., R.D.: designed and performed the experiments, and wrote the manuscript. M.J., A.P., A.M.: analysed the data; D.B. was involved in the clinical management of the patient. All authors contributed and approved the final version of the manuscript.
Conflict of interest
The authors declare no competing financial interest.
Acknowledgements
This work was supported by institutional funds and Department of Biotechnology, Ministry of Science & Technology, New Delhi (BT/PR12682/MED/12/689/2015). M.J. and A.A. received a fellowship from the Indian Council of Medical Research (ICMR), New Delhi. Authors show gratitude to all the technical and clinical staff involved in the patient management and laboratory diagnosis.




