Diffuse large B cell lymphoma cell of origin by digital expression profiling in the REAL07 Phase 1–2 study
Diffuse large B-cell lymphoma (DLBCL) has been molecularly classified in two main subtypes of cell of origin (COO) profile reflecting different stages of B-cell differentiation: germinal centre B-cell (GCB) from germinal centre (GC) centroblasts, and activated B-cell DLBCL (ABC) from GC cells undergoing plasmacytic differentiation (Testoni et al, 2015). The sub-classification of DLBCL based on gene expression profiling (GEP) has not only proved to be a landmark in understanding the pathogenesis of the disease but it also has a clinical relevance since the two subtypes present different outcome with the standard R-CHOP regimens (Testoni et al, 2015). A series of immunohistochemistry (IHC)-based approaches have been proposed as alternatives, but without fully recapitulating the GEP results (Testoni et al, 2015). Two groups have designed digital expression profiling assays to determine the DLBCL COO on formalin-fixed paraffin-embedded (FFPE) tissue (Masque-Soler et al, 2013; Scott et al, 2015). Both assays consist of a 20-gene panel chosen after an extensive data mining procedure of public available data (Masque-Soler et al, 2013; Scott et al, 2015). Here, a zero-sum implementation (Altenbuchinger et al, 2017) of the signature published by Masque-Soler (Masque-Soler et al, 2013) was applied in a cohort of 47 DLBCL cases selected from the 61 patients enrolled into the REAL07 phase 1b/2 study (Chiappella et al, 2013; Vitolo et al, 2014); the selection of cases was based on the availability of material. The REAL07 was a multicentre trial conducted by the Fondazione Italiana Linfomi, assessing the maximum tolerated dose (MTD), the safety and efficacy of lenalidomide in addition to standard chemo-immunotherapy rituximab plus cyclophosphamide-doxorubicin-vincristine-prednisone (R-CHOP) given every 21 days in elderly DLBCL patients. Eligibility, exclusion criteria and clinical results, including the COO as obtained using the Hans IHC algorithm (Hans et al, 2004) of the 47 patients treated at the MTD of the phase 2 part of the study, have been previously described (Vitolo et al, 2014). The nucleic acids were extracted using the Qiagen AllPrep FFPET kit and RNA was quantitated using spectrophotometry (NanoDrop, Thermo Scientific, Wilmington, DE, USA): 47/47 (100%) samples passed the quality control and underwent digital GEP on 200 ng RNA using NanoString technology. The application of the Masque-Soler panel in 47 samples led to the identification of 12 ABC (26%), 17 GCB (36%) and 18 unclassified cases (38%) (Fig 1). When we compared the Nanostring-based approach and the IHC method, 36 cases were designated as non-GCB or GCB by the IHC, the assay differed in the classification of two cases: one ABC case was assigned to GCB group and one GCB case was assigned to ABC group (6% rates of miss-assignments). When considering the whole dataset including 11 unclassified cases, the assay classified three GCB patients as unclassified, 10 ABC patients as unclassified and the 11 unclassified patients as two ABC, five unclassified and four GCB. Outcomes of the 47 patients were used to determine whether the COO assignments made by the assay maintained the prognostic significance. Differently from that seen with R-CHOP based regimens (Testoni et al, 2015) and in line with the results presented in the original REAL07 phase 1-2 study publication (Vitolo et al, 2014), the ABC groups defined by the assay did not show a worse progression-free survival than the GCB group (P > 0·05). Our results underline that the similar outcome in GCB and non-GCB patients could not be due to a random misclassification from the IHC analysis because we used a GEP-based approach. However, it would be advisable to directly compare different available approaches to define the COO (Masque-Soler et al, 2013; Scott et al, 2015; Gleeson et al, 2016; Reinholz et al, 2016), possibly also investigating additional features, such as MYC or BCL2 deregulation, in a single assay (Testoni et al, 2015).
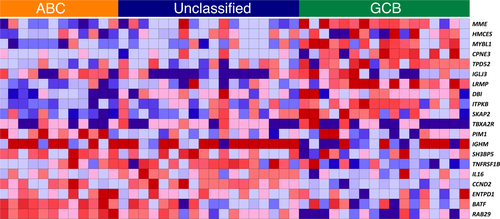
Here, the number of unclassified patients was higher than originally described using the Hans IHC algorithm (Vitolo et al, 2014) or compared to what reported by Scott et al (2015) using the other Nanostring-based assay, but it was consistent with other data obtained with microarray-based GEP on FFPE samples (Gleeson et al, 2016). Modifications of data mining approaches can certainly reduce this number but it would be important to properly define the biological and the clinical relevance of the misclassified cases, especially when specific therapies will be approved for an individual COO-defined DLBCL subtype.
In conclusion, these results acknowledge that the regimen of lenalidomide plus R-CHOP21 is potentially efficacious in the ABC patients, as also very recently reported in another R2CHOP phase 2 study (Nowakowski et al, 2015). Although in a small cohort of patients, the assay reported by Masque-Soler et al (2013) could be applied to all FFPE samples collected in the context of a prospective clinical trial and was able to assign the COO.
Authorship contribution
LC performed data mining, interpreted data and co-wrote the manuscript; AR, performed experiments; AC, UV provided clinical data; IK, GC, MA, CK performed data mining; GI characterized and provided samples, co-designed the study; FB co-designed the study, interpreted data and co-wrote the manuscript. All authors have read and approved the manuscript.
Acknowledgements
Work partially supported by the Barletta and the Gelu Foundations. We thank our Colleague Wolfram Klapper for kindly providing access to the probes design for the Nanostring assay and for the helpful discussion.
Conflict of interest
AC, UV, GI have received research funds from Celgene. The other authors have no conflicts of interest.




