Factors predicting the identification of new organisms in follow-up blood cultures drawn during episodes of neutropenic sepsis
While patients presenting with neutropenic fever have high rates of initial bacteraemia (Freifeld et al, 2011), follow-up blood cultures appear to have a very low yield (Serody et al, 2000; Wattier et al, 2015; Neemann et al, 2016). As previous studies have examined only small numbers of patients, we used a data linkage approach to assess the frequency and predictors of new isolates in follow-up blood cultures during episodes of suspected neutropenic sepsis.
Blood cultures drawn during episodes of suspected neutropenic sepsis in haematology patients at St Vincent's Hospital Melbourne between January 2008 and June 2016 were identified by linking data from multiple institutional databases as outlined in the online Data S1. The protocol was approved as a quality-assurance activity by our Quality and Risk unit.
The clinical haematology unit at our institution admits 450 patients and performs 20–25 autologous haematopoetic stem cell transplants per year. Approximately a quarter of patients screened on the haematology ward are found to be colonised with vancomycin-resistant Enterococcus faecium (VRE), and the ward was affected by an institutional outbreak of KPC2-producing Klebsiella pneumoniae during the study period (Cronin et al, 2017). Throughout the study period, institutional guidelines recommended piperacillin-tazobactam monotherapy as the first-line empiric regimen with vancomycin added for severe sepsis and suspected central line infection. Primary fluoroquinolone prophylaxis was not used during the study period.
Clinical data and blood culture results were extracted from institutional databases as outlined in the supplementary materials. Isolates were regarded as contaminants if this was documented by medical microbiology staff at the time of the original result using the national surveillance definition (Australian Commission on Safety and Quality in Health Care 2010). New isolates differed in their identification (to species-level) and/or their antimicrobial susceptibility pattern from all other preceding isolates in an episode.
Binomial logistic generalised estimating equations with an exchangeable correlation structure were used to evaluate seven clinical predictors of a new isolate in blood cultures collected more than 24 h after the first blood culture of an episode (‘follow-up blood cultures’). Further details regarding the statistical analysis are in the Data S1. Variables with P < 0·20 on univariate analysis were included in a multivariate model where statistical significance was defined as P < 0·05. All statistical analysis was performed using Stata v12·1 (StataCorp, College Station, TX, USA).
During the study period, 91 767 blood cultures were collected from 26 715 patients at our institution. After linkage, 6881 were organized into 912 episodes in 502 haematology inpatients. The most common diagnosis in this series was acute myeloid leukaemia (AML, 315 episodes). The absolute neutrophil count (ANC) on the day of the first blood culture was <0·1 × 109/l in 571 episodes (63%), and 469 episodes (51%) occurred in patients known to be colonised with vancomycin-resistant enterococci (VRE) prior to the start of the episode. Additional descriptive information can be found in Tables SI and SII.
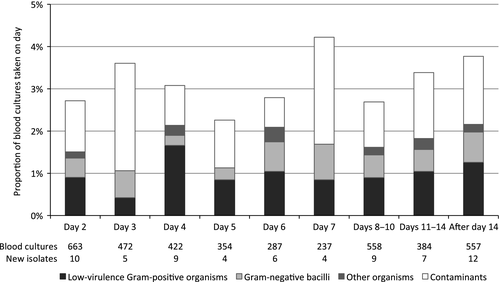
A total of 3933 follow-up blood cultures were drawn in 602 episodes from 376 patients: 3611 (92%) were negative, 200 (5%) grew isolates previously identified during the episode, 66 (1·7%) grew new isolates, and 57 (1·4%) grew contaminants. Only 361 (9%) were the first negative blood culture after a previous positive (establishing ‘clearance’ of bacteraemia). The majority of new isolates in follow-up cultures were low-virulence gram-positive organisms (Enterococcus faecium 26, coagulase-negative staphylococci 10, Corynebacterium jeikeium 1, E. faecalis 1, E. raffinosus 1). Nineteen of the new E. faecium isolates were vancomycin-resistant, most of which were from patients with previously-documented colonisation (16 cases). Central line infections were the source of 13 new isolates. The proportion of cultures growing new isolates and the distribution of organism types were stable over time (Fig 1).
Neither VRE colonisation (P = 0·90) nor a previous positive blood culture (P = 0·69) predicted a new isolate on univariate analysis (Table SIII). The other five variables were entered into the multivariate analysis (Table 1). The area under the receiver-operating-characteristic curve (ROC AUC) for a model using the four variables with P < 0·05 was 0·75 (asymptotic 95% confidence interval 0·69–0·81).
| Variable | Blood cultures (n = 3933) | New isolates (n = 66) | OR | 95% CI | P |
|---|---|---|---|---|---|
| Acute myeloid leukaemia | 1936 | 50 | 2·21 | 1·34–3·66 | 0·002 |
| ANC <0·1 × 109/l | 2311 | 57 | 3·70 | 1·77–7·73 | 0·001 |
| Days of ANC <0·1 × 109/l preceding collection | – | – | 1.01 | 0·99–1·02 | 0·30 |
| Time since preceding blood culture (per day) | – | – | 1·29 | 1·13–1·48 | <0·001 |
| ICU admission | |||||
| ≥12 h prior to ICU admission, if any | 3298 | 47 | Base | ||
| <12 h prior to admission to ICU | 30 | 3 | 7·53 | 2·09–27·2 | 0·002 |
| During an ICU admission | 199 | 8 | 3·20 | 1·58–6·49 | 0·001 |
| After discharge from ICU | 406 | 8 | 1·18 | 0·62–2·25 | 0·61 |
- ANC, absolute neutrophil count; CI, confidence interval; ICU, intensive care unit; OR, odds ratio.
Only seven (0·6%) of the 1527 follow-up blood cultures (39% of all follow-up blood cultures) taken from patients with an ANC ≥0·1 × 109/l who were not in the intensive care unit (ICU) were positive for a new isolate. Six of these were low virulence gram-positive organisms (E. faecium 3, coagulase-negative staphylococci 3), and one was due to a S. aureus central line infection. None of these seven patients (one isolate each) were subsequently admitted to the ICU or died in hospital. The only other new isolates from patients with an ANC ≥0·1 × 109/l were two different Enterobacteriaceae grown on one day from a single ICU patient who died 4 days later despite pre-existing antibiotic therapy to which the isolates were sensitive in vitro. New isolates were also uncommon in cultures drawn when the ANC was <0·1 × 109/l, particularly in patients with diagnosis other than AML (9 of 901, 1%). This association with AML did not seem to relate to the preceding duration of profound neutropenia.
Blood cultures taken within 48 h of the previous set (“daily” blood cultures) were less likely to grow a new isolate (47 of 3465, 1·4%) than those taken after a longer interval (19 of 462, 4·1%). This finding, and the association with ICU admission, suggests that clinical stability (which might include ongoing fever) argues against new-onset bacteraemia.
This study was limited by the data accessible for linkage at our institution – the presence of fever, central access devices and antibiotic therapy at the time of each blood culture would have been variables of interest. We did not assess the association between C-reactive protein levels and the demonstration of a new isolate as this test is not routinely repeated on a daily basis at our institution, complicating the assignment of a particular value to each blood culture and preventing the consistent analysis of trends. In addition, the very low rate of new isolates and the presence of predictive variables among data collected as part of routine haematological care would not seem to support the adoption of an additional routine test for the purpose of triaging patients for repeat blood cultures. The lack of an allogeneic haematopoetic stem cell transplantation programme at our institution prevented us from examining this particularly high-risk population.
In conclusion, follow-up blood cultures in patients with suspected neutropenic sepsis have a low yield for new organisms, particularly for patients with an ANC ≥0·1 × 109/l or diagnoses other than AML. Clinicians should feel comfortable not performing repeat blood cultures unless there is a significant change in a patient's condition or a specific suspicion of new-onset bacteraemia.
Acknowledgements
The authors would like to acknowledge Prof Allen Cheng for his statistical advice.
Authorship contribution
GSH designed the study, performed the research, analysed the data and wrote the paper; CT and MJW interpreted the data and wrote the paper.
Competing interests
The authors have no competing interests.




