Single-nucleotide variations defining previously unreported ADAMTS13 haplotypes are associated with differential expression and activity of the VWF-cleaving protease in a Salvadoran congenital thrombotic thrombocytopenic purpura family
Although autosomal recessive haematological disorders, such as congenital thrombotic thrombocytopenic purpura (cTTP), are individually rare and difficult to ascertain, studies involving one or more homozygous affected children and their unaffected heterozygous parents have led to expanded understanding of known and discovery of previously unknown molecular-genetic characteristics. We present an in-depth examination of ADAMTS13 haplotypes, mRNA levels and protein expression, activity, and enzyme kinetics in this case study of two Salvadoran children with cTTP – the first reported cases of this disease in individuals originally from Central America – and their parents.
The propositi – a son and a daughter born to asymptomatic, non-consanguineous (but possibly distantly related) parents from the same town in El Salvador – were diagnosed with cTTP at ages 6 and 2 years, respectively. Although they developed haemolytic anaemia and thrombocytopenia 2 d (son) and 17 months (daughter) after birth, their cTTP diagnosis was not rendered until both were hospitalized simultaneously with fever, respiratory symptoms, haemolytic anaemia and thrombocytopenia. Peripheral blood smears for both demonstrated schistocytes, and their von Willebrand factor-cleaving protease (VWF-CPase) activities were <1% without ADAMTS13 IgG antibodies. Fresh frozen plasma (FFP) infusion induced rapid (within 48 h) normalization of their blood counts and resolution of the microangiopathic changes. Both children have been treated with prophylactic infusions of FFP (10 ml/kg every 2·5 weeks) without long-term neurological or renal sequelae.
Following approval by institutional review boards and receipt of informed consent, blood samples were collected from the children, prior to FFP administration, and from both parents. Genomic DNA and total RNA were extracted from peripheral blood leukocytes using QIAamp DNA Blood Maxi and PAXgene Blood RNA kits, respectively (Qiagen; Germantown, MD, USA). All ADAMTS13 exons, at least 50 base pairs (bp) of each flanking intron junction, ~500 bp 5' of the promoter, and ~200 bp of 3'-flanking genomic DNA were amplified using polymerase chain reaction (PCR), cleaned using Exo-SAP-IT (Affymetrix; Santa Clara, CA, USA), and subjected to direct bi-directional Sanger sequencing followed by capillary electrophoresis and analysis on ABI-3730 instruments. Reverse transcription was performed (Shomron et al, 2010), and plasma VWF-CPase antigen was measured using the Technozym ADAMTS13 enzyme-linked immunosorbent assay (Technoclone; Vienna, Austria) and the Victor X3 multilabel plate reader (PerkinElmer; Waltham, MA, USA). Fluorogenic FRETS-VWF73 (Peptides International; Louisville, KY, USA) was prepared and assayed (Sauna et al, 2009). The kinetic characteristics of ADAMTS13 were obtained using GraphPad Prism software.
ADAMTS13 sequencing revealed both children to be homozygous and their parents to be heterozygous for the previously described, cTTP-causing, single-base-substitution mutation 20506C > T (Table 1A) (Levy et al, 2001; Snider et al, 2004; Hing et al, 2013). The children's VWF-CPase antigen and activity levels were undetectable, although steady-state levels of the ADAMTS13 mRNA were >2·5-fold higher in the daughter than in the son. The re-sequenced regions of the ADAMTS13 loci segregating within this family contained 26 additional single-nucleotide variations (SNVs), including two previously unreported non-synonymous (ns)-SNVs: 27852C > T (c.3362C > T ⇒ 972Arg > Trp) and 33325G > A (c.3733G > A ⇒ 1096Arg > His) (Table 1A). The parents' genotypes differed at nine positions, including three ns-SNVs, creating two distinct, non-mutant haplotypes (designated I and III) at the gene, mRNA, and protein levels (Table 1A). The quantitative real-time-PCR assay revealed >4-fold higher steady-state mRNA levels in the father compared to the mother (P < 0·001; Table 1B). Plasma VWF-CPase activity and antigen levels were ~2-fold greater in the father than in the mother (P = 0·00164 and P = 0·0633, respectively), but the specific activities of these structurally distinct VWF-CPase proteins were notably almost identical (253·5 vs. 256·2 U/μg). Initial velocity kinetic analysis using the Michelis-Menten equation demonstrated that the Vmax of the father's VWF-CPase was twice that of the mother's (1·4 vs. 0·7; P < 0·0001) while its affinity for substrate (FRETS-VWF73) was one-third that of her VWF-CPase (Km = 0·3 vs. 0·1; P = 0·0585).
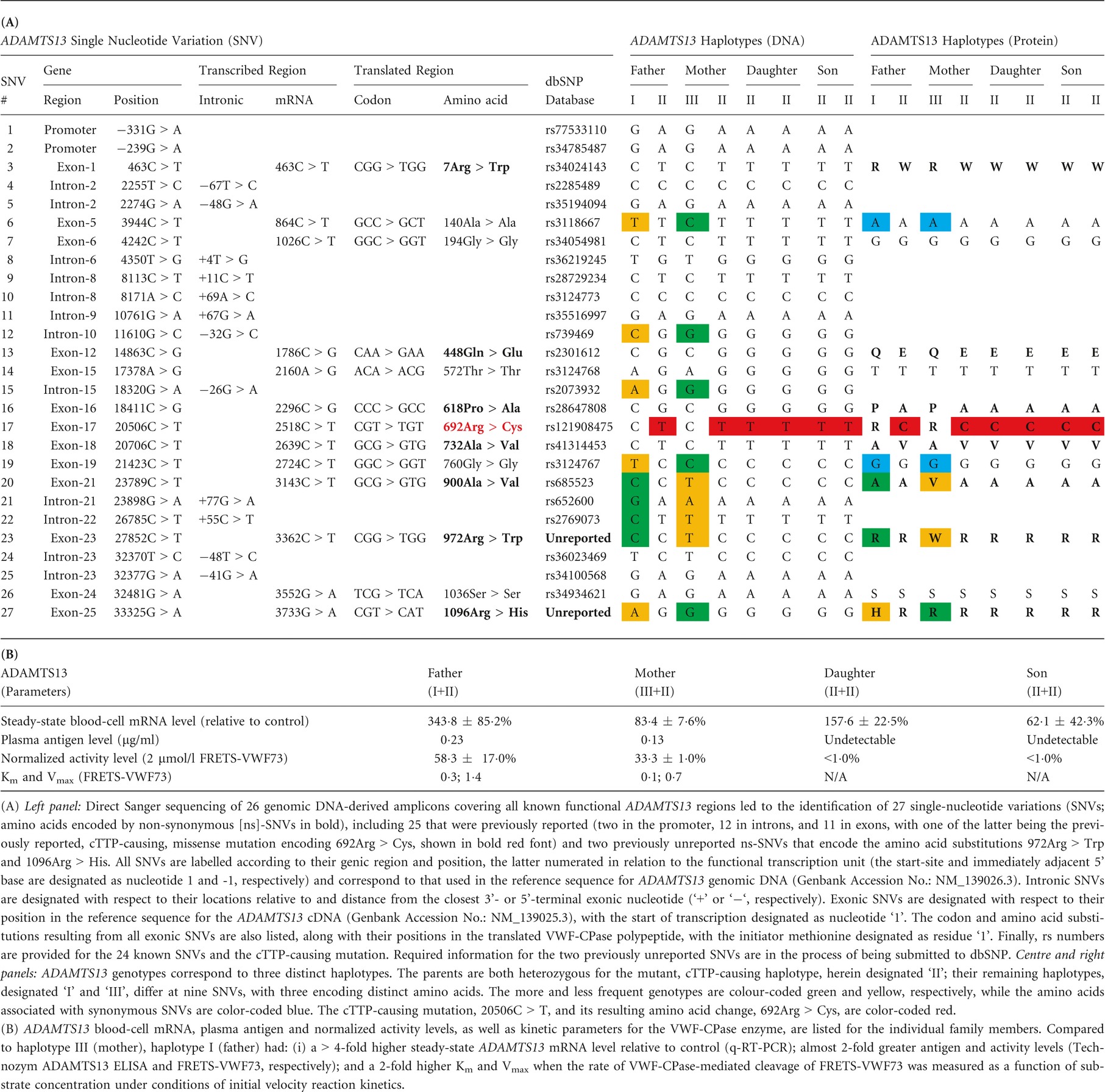
The absence of any detectable VWF-CPase antigen or activity in the affected children was consistent with findings from a previous study, which reported that the resultant amino acid substitution (20506C > T, 692R > C) probably prevents its secretion due to endoplasmic reticulum retention (Meyer et al, 2008). As a consequence of this impaired secretion, we were able to associate the measured circulating VWF-CPase antigen and activity levels observed in the father and mother with their distinct, non-mutant ADAMTS13 alleles.
While prior studies have shown that the synonymous (s)- and ns-SNVs 3944C > T (c.864C > T ⇒ 140Ala > Ala) and 23789C > T (c.3143C > T ⇒ 900Ala > Val), respectively, are associated in vitro with no or only minor effects on VWF-CPase secretion compared to their more frequent wild-type alleles (Plaimauer et al, 2006; Edwards et al, 2012), the seven other allelically distinct, parental ADAMTS13 potential quantitative-trait nucleotides (QTNs) have never been experimentally investigated, individually or in combination with each other. The candidate functional SNVs, 11610G > C (IVS10[-32G > C]) and 18320G > A (IVS15[-26G > A]), are located in intronic regions known to contain cis-elements comprising 3'-splice junctions, and their alleles may, for example, influence precursor mRNA processing differentially. The two s-SNVs, 3944 C > T (c.864C > T ⇒ 140Ala > Ala) and 21423C > T (c.2724C > T ⇒ 760Gly > Gly), are also candidate QTNs, as the alleles of such variants have been reported to influence differentially the rates of translation, post-translational modification and/or secretion, all of which can affect the structure and/or function of the encoded polypeptide (Sauna & Kimchi-Sarfaty, 2011). Finally, the three ns-SNVs – 23789C > T (c.3143C > T ⇒ 900Ala > Val), c.3362C > T (972Arg > Trp), and c.3733G > A (1096Arg > His) – are obvious potential QTNs as changes in the VWF-CPase primary structure may modulate its function directly. Additionally, these SNVs are linked across the non-mutant ADAMTS13 haplotypes, I and III, and they may influence gene expression and/or function in a more complex, combinatorial manner as a quantitative trait locus. Although this study suggests that ADAMTS13 may be a major determinant of inter-individual variability in the expression and/or function of the VWF-CPase, recent findings did not reveal significant genetic linkage between microsatellites within or near the encoding structural locus for ADAMTS13 in chromosome 9 (Viel et al, 2007). While beyond the scope of this letter, in vitro expression studies involving each SNV separately or in combination will ultimately be necessary to determine which of the myriad potential mechanisms are responsible for the observed differences in circulating VWF-CPase antigen and activity in vivo and improve our discernment of ADAMTS13 genotype-phenotype relationships.
Acknowledgements
The authors thank Dr. Shelley Cole (Texas Biomedical Research Institute; San Antonio, Texas) as well as Ms. Uma Dandekar and her staff at the UCLA GenoSeq genomics core facility for their help with DNA sequencing. We also thank Dr. Kevin Viel (Histonis, Inc.), Ms. Minghong Ward (NCBI, NIH) and Mr. Brian Anderson (DNASTAR, Inc.) for their help with DNA sequence analysis. Research conducted in the laboratory of CKS is funded by the Division of Hematology, Center for Biologics Evaluation and Research, Food and Drug Administration. Research conducted in the laboratory of TEH is funded by grants from the National Heart, Lung and Blood Institute, NIH (1RC2-HL101851, HL-71130 and HL-72533), as well as from the Bayer Healthcare Corporation, Bayer Hemophilia Awards Program, Baxter Healthcare Corporation, and the Clinical Translational Science Institute at the University of Southern California.
Authors' contributions
TEH and CKS designed the research study. BK, ZAH, AW, TS, EBS, DL, NCE, and VLS performed the research. EJB, PHK, VJM, and ZES contributed essential reagents or tools. BK, TEH, ZAH, and CKS analysed the data. BK, AW and ZAH wrote the paper.
Disclaimer
The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any Agency determination or policy.




