Fusion of the additional sex combs like 1 and teashirt zinc finger homeobox 2 genes resulting from ider(20q) aberration in a patient with myelodysplastic syndrome
A variant of del(20q), an isochromosome of the long arm with the loss of an interstitial part of 20q, ider(20q), has been reported in patients with myeloid diseases (Li et al, 2004). About 40 cases with this rearrangement have been reported up to 2012 (reviewed by Mullier et al, 2012). Molecular cytogenetic and array techniques have been used for mapping of the deleted region on 20q (Douet-Guilbert et al, 2009). The proximal breakpoints are consistently located in the 20q11.21 band, and the distal breakpoints span from band 20q13.13 to band 20q13.33.
A 79-year-old male patient was admitted to our hospital with a diagnosis of refractory anaemia with ring sideroblasts (RARS). His haemoglobin level was reduced to 108 g/l; other blood parameters were normal. A bone-marrow aspirate was typical for RARS and revealed mild hypogranular and strong vacuolated neutrophils. No specific haematological therapy was suggested, other than transfusion-supported treatment. The patient provided his written informed consent for the use of his sample for research purposes.
The 46,XY,ider(20)(q10)del(20)(q11q13) karyotype was established in bone marrow cells. Fluorescence in situ hybridization (FISH) with a locus-specific probe, ON MDS 20q- (PTPRT 20q12)/20q11 (KREATECH Diagnostics, Amsterdam, the Netherlands) confirmed the 20q11/20q12 deletion and FISH studies with a subtelomeric probe, Vysis ToTel 20p/20q (Abbott Molecular, Des Plaines, IL, USA), proved the duplication of the subtelomeric 20q region and the deletion of the subtelomeric 20p region. Multicolour FISH banding of chromosome 20 (XCyte 20; MetaSystems, Altlussheim, Germany) showed ider(20)(q10)del(20)(q11.1q13.1). Array comparative genomic hybridization (aCGH, CytoChip Cancer 180K, BlueGnome, Cambridge, UK) was used to detect gene copy number variations. Two duplicated regions, the first ranging from 20p11 to 20q11.21 (bp 25 805 264–30 960 195) and the second from 20q13.2 to 20q13.33 (bp 52 080 063–62 949 120), were detected with two other deletions that included the short arm of chromosome 20 (except 20cen-20p11.2) and the 20q11.21–20q13.2 region (bp 30 971 874–52 045 077) (Fig 1A). This analysis confirmed that ider(20q) was a dicentric chromosome. Metaphase FISH mapping with a set of six bacterial artificial chromosome (BAC) probes (BlueGnome) distributed in 20q11.21 and 20q13.2, with a chromosome-20-specific centromeric probe [SE 20 (D20Z1); KREATECH Diagnostics] was performed. Based on the results (Table 1), we deduced that the region 20q11.21–20q13.2, including parts of the additional sex combs like 1 (ASXL1) and teashirt zinc finger homeobox 2 (TSHZ2) genes, had been deleted and the corresponding RNA levels were analysed to confirm this hypothesis. RNA was isolated from mononuclear cells using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and cDNA was prepared (Marková et al, 2009). Four forward primers covering the hypothetically disrupted 5′ end of the ASXL1 gene were designed, together with 15 reverse primers distributed along the whole TSHZ2 gene. Eight multiplex polymerase chain reaction (PCR) reactions were performed using one of the forward primers together with a mixture of reverse primers 1–8 or 9–15. The amplified products were subjected to electrophoresis on 2% agarose gel and the only band amplified (247 bp, resulting from the ASXL1-for2 and TSHZ2-rev1–8 mixed primers) was directly sequenced using the forward primer and the BigDye Terminator Cycle Sequencing Kit v. 3.1 (Applied Biosystems, Branchburg, NJ, USA). Sequencing analysis revealed a fusion between exon 4 of the ASXL1 gene and exon 3 of the TSHZ2 gene. This result was validated with a single PCR using primers ASXL1-for2 (5′-GTCATAGAGGCAGAAGGACT-3′) and TSHZ2-rev5 (5′-AGGAGGAGTTCAATGAGTTC-3′).
| Locus | BACs | Size (bp) | Signal on ider(20q) |
|---|---|---|---|
| p13a | Subtelomere 20p | − | |
| p10-q10b | ++ | ||
| q11.21 | RP1-316I5 | 30 898 114–30 994 333 | ++ |
| q11.21 | RP11-358N2 | 30 938 520–31 115 717 | ±± |
| q11.21 | RP5-1184F4 | 31 0231 30–31 141 902 | − |
| q11/q12c | −/− | ||
| q13.2 | RP4-669H2 | 51 901 848–52 021 567 | − |
| q13.2 | RP5-823G15 | 51 979 824–52 146 749 | ++ |
| q13.2 | RP4-724E16 | 52 128 204–52 255 974 | ++ |
| q13.3a | Subtelomere 20q | ++ |
- − (−/−), signal(s) missed; ++, two signals present; ±±, two weak signals present.
- a ToTel 20p/20q (Abbott Molecular, Des Plaines, IL, USA).
- b SE 20 (D20Z1) (KREATECH Diagnostics, Amsterdam, the Netherlands).
- c ON MDS 20q- (PTPRT 20q12)/20q11 (KREATECH Diagnostics).
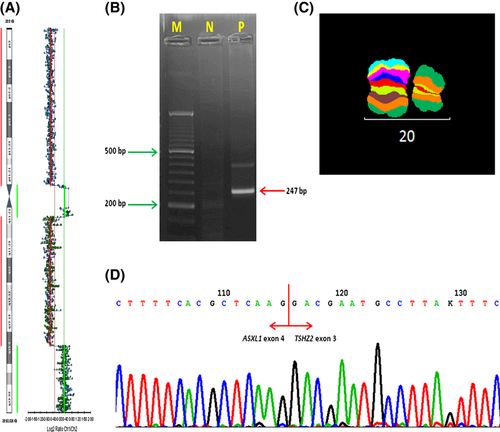
Using a combination of all the techniques described above, the karyotype was shown to be 46,XY,idic(20)(p11)del(20)(q11.21q13.2), with the fusion gene ASXL1/TSHZ2 (Fig 1).
The frequency of ider(20q) is very low, this is the first case we have observed in approximately 1500 patients with myeloid malignancies examined during the last 10 years. The identification depends on the resolution of the methods used. Li et al (2004) used G- and R- banding techniques in combination with FISH to first describe this aberration as ider(20)(q10)del(20)(q11q13). In follow-up studies, the same group revised all previously observed cases by mapping the 20p11.21–20p11.22 region with a BAC/PAC contig and showed that ider(20q) is a dicentric chromosome (Li et al, 2006). We delimited the proximal and distal breakpoints in our patient using aCGH and FISH with sets of BAC probes for the 20q11.21 and 20q13.2 regions respectively, and confirmed the aberrant dicentric chromosome. On the mRNA level, the disruption and partial loss of the ASXL1 (exons 5–12) and TSHZ2 (exons 1 and 2) genes was described. The ASXL1 gene maps to chromosomal region 20q11 and belongs to a family of three paralogues. This gene is one of the most frequently mutated genes in malignant myeloid diseases, and the majority of mutations affect exon 12 (with the consequent loss of the PHD domain), although a few deletions and translocations with fusions to the PAX5 gene have also been reported (reviewed by Gelsi-Boyer et al, 2012). Recent studies have also highlighted the role of the truncation of the PHD domain in malignant progression (Baker et al, 2008). The TSHZ2 gene is located in the 20q13.2 band and its role in aberrant methylation has been demonstrated in breast and prostate cancer, when the TSHZ2 and TSHZ3 genes were shown to be the most important candidates for novel tumour suppressor genes (Yamamoto et al, 2011). The TSHZ2 gene was partially deleted in our patient, so we hypothesized that its expression was downregulated. Similarly, the DNMT3B gene, responsible for DNA methylation, and MYBL2, a tumour suppressor gene, were deleted, which may contribute to the malignant transformation of cells.
Several mechanisms accompanying the formation of ider(20q) contribute to the tumourigenesis of myeloid malignancies—the loss of tumour suppressor genes in the deleted regions on the short and long arms of chromosome 20 (Clarke et al, 2013); the overexpression of genes in the duplicated regions (Mackinnon et al, 2010); the aberrant methylation/demethylation of gene promoters (Yamamoto et al, 2011); and the formation of fusion genes. To our knowledge, this is the first report describing ASXL1/TSHZ2 fusion gene. The identification of further cases will be necessary to determine the frequency and significance of the ASXL1/TSHZ2 fusion and its implications for the treatment of patients with this rare but recurrent aberration.
Acknowledgements
J.B. was responsible for the design of the study, the interpretation of the molecular cytogenetic results, and with H.B., for writing the manuscript. I.S., S.R., and S.I. performed the FISH analyses, and participated in the design and interpretation of the experiments. H.B. and K.K. performed the aCGH, and Z.Z. was responsible for the interpretation of the results of these two techniques and participated in the revision of the manuscript. J.M. performed the RNA and PCR analyses. J.S. provided medical care for the patient. K.M. supervised the manuscript preparation. All the authors read and approved the final manuscript.
Funding
This work was supported by grants UHKT2005 00023736, RVO-VFN64165, GACR P302/12/G157/1, and PRVOUK-P27/LF1/1.
Conflict of interest
All authors disclose that they have no conflicts of interest.




