Genotypic diversity of Pasteurella multocida isolates from pigs and poultry in Australia
Abstract
Objective
To investigate the genotype and diversity of Pasteurella multocida present in pig herds and to determine the extent of overlap with isolates from poultry flocks in Australia.
Methods
A total of 43 isolates from pigs from different farms and regions of Australia were used in this study. A diverse collection of 41 poultry isolates, with 31 being previously characterised, was also used. The pig isolates and 10 poultry isolates were identified by species-specific PCR assay, serotyped by the Heddleston scheme and genotyped by a multiplex PCR based on the lipopolysaccharide (LPS) outer core biosynthesis locus, repetitive element PCR fingerprinting (rep-PCR) and multilocus sequence typing (MLST), with the latter being used on a subset of the isolates based on the rep-PCR results.
Results
Only 4 out of 8 recognised LPS genotypes were found in the pig isolates, with each isolate assigned to an LPS genotype. In contrast, 77% of the isolates were non-typable or cross-reacting in the Heddleston serotyping scheme. The rep-PCR analysis recognised 20 patterns, yet only 16 sequence types (STs) were found and 4 were new STs. There were 5 STs (STs 7, 11, 20, 24 and 58) shared among the pig and poultry isolates.
Conclusions
Although only limited numbers of isolates have been examined, there is evidence of a sharing of genotypes between Australian pigs and chickens. These findings have major implication for biosecurity measures with regard to minimising both direct (e.g. animal to animal) and in-direct (e.g. shared staff or cross-visitors) contact between poultry and pigs.
Abbreviations
-
- LPS
-
- lipopolysaccharide
-
- MLST
-
- multilocus sequence typing
-
- NT
-
- non-typable
-
- rep-PCR
-
- repetitive element PCR fingerprinting
-
- RIRDC
-
- Rural Industries Research & Development Corporation
-
- ST
-
- sequence type
Pasteurella multocida belongs to the family Pasteurellaceae, a family whose members are among some of the main causes of highly infectious diseases that negatively affect livestock industries.1 In poultry, P. multocida causes fowl cholera, which is a serious disease with clinical signs that include depression, ruffled feathers, fever, anorexia, mucous discharge from the mouth, diarrhoea, increased respiratory rate, septicaemia and death.2 In pigs, P. multocida is regarded as an opportunistic pathogen and is associated with the pig respiratory disease complex.3 This co-infection status makes it hard to attribute symptoms in pigs associated with P. multocida, because the clinical signs and lesions are normally superimposed on that of the primary agent.3 Recent challenge studies have shed light on the symptoms of pneumonia in pigs, with dyspnoea and hyperthermia being the main clinical symptoms.4 As well, P. multocida in pigs is associated with progressive atrophic rhinitis, with obvious symptoms including shortening and twisting of the snout.1
In the Australian context, somatic Heddleston serotyping, lipopolysaccharide (LPS) genotyping and repetitive element PCR fingerprinting (rep-PCR) are being used to guide autogenous vaccine use in the poultry industry. However, characterisation studies on Australian pig isolates have been very limited since studies in the 1990s.5-8
It is known that P. multocida isolates of the same genotype can be obtained from cats and chickens on the same farm9 and it has been shown that isolates from pigs are pathogenic for poultry.10 However, even free-range chickens in close contact with other animal species in a village in Tanzania did not show a wide exchange of P. multocida between species.11 The diversity of poultry P. multocida isolates in Australia has been previously assessed by phenotypic12 and genotypic13 methods. Similarly, the diversity of Australian pig isolates has been examined in terms of phenotypic properties,5 as well as by genotypic methods such as restriction endonuclease analysis, ribotyping and plasmid analysis.6-8 Multilocus sequence typing (MLST)14 has proven useful in two Australian-based studies looking at fowl cholera outbreaks.9, 15
The Rural Industries Research & Development Corporation (RIRDC) was set up as an MLST scheme originally based on poultry isolates and the Heddleston serovar reference strains.16 The RIRDC P. multocida MLST has recently been combined with an alternative scheme (available at the PUBMLST website: http://pubmlst.org/pmultocida/) and both databases aim to investigate the evolutionary relationships between bovine, ovine, porcine and avian isolates of P. multocida.
In the current study, we examined a collection of Australian pig and poultry P. multocida isolates using conventional Heddleston serotyping, LPS genotyping and MLST. We used the results to evaluate the diversity of these isolates and then compared the results with those from prior poultry studies9, 15 to look for evidence of overlap of P. multocida genotypes across the two host species using the MLST databases.
Materials and methods
Bacteria
This study utilised 43 isolates from pigs (Supplementary Table 1) and 41 isolates from poultry (Supplementary Table 2) collected as part of the Department of Agriculture and Fisheries/Queensland Queensland Alliance for Agriculture and Food Innovation reference diagnostic services.
The pig isolates were collected from the lung, brain or peritoneal fluid of diseased animals. Isolates were deliberately selected for diversity, being from 33 different farms (Farms 1–33) across regions of Australia. The isolates were identified, serotyped via the Heddleston typing scheme and genotyped by the LPS multiplex PCR. A rep-PCR method, described later, was used to determine the different strains among the pig isolates, with representative isolates then being genotyped by the MLST scheme.
The chicken isolates were isolated from pericardial sac, heart blood, liver or bone marrow of the femur. Among the 41 field isolates from poultry, 31 came from previous studies9, 16, 17 and had been serotyped and genotyped by MLST. As well, 21 of these 31 isolates had been examined by LPS multiplex PCR.17 Hence, a total of 20 isolates were analysed by LPS multiplex PCR for the purpose of this study only. A subset of 10 isolates was also identified, serotyped by the Heddleston scheme and genotyped by MLST.
Most of the isolates were sent on blood agar plates or slopes from frontline laboratories; some were collected on farm via swabs and transported to the laboratory on ice before being plated on 5% sheep blood agar and incubated in air at 37°C overnight.
Identification and serotyping of P. multocida
The P. multocida isolates were identified by a species-specific PCR18 and then serotyped with the Heddleston serotyping scheme as described previously.19
The LPS multiplex PCR, which targets the LPS outer core biosynthesis locus, was performed as previously described16 and assigned isolates to one of the eight LPS types (termed L1–L8).16 The genomic DNA for this PCR was prepared using the PrepMan Ultra Sample Preparation Reagent according to manufacturer’s protocol (Life Technology; Applied Biosystems, Warrington, UK).
Genotyping by rep-PCR
A rep-PCR assay was performed as previously described20 using template prepared by the PrepMan Ultra kit. Commercial software (Bionumeric version 4.50, Applied Maths Inc., Saint-Martens-Latem, Belgium) was used to analyse the patterns. If two isolates had the same genomic fingerprint (i.e. an identical band pattern including size and intensity), they were assumed to be the same strain. This assumption was based on the original study developing the RIRDC MLST scheme for P. multocida, which suggested that the MLST data matched the results given by rep-PCR.16 Within each rep-PCR genotype found in the pig isolates, a representative isolate was subjected to MLST.
Genotyping by MLST
MLST genotyping was based on the RIRDC MLST scheme of sequencing 466–602 base-pair internal fragments of seven housekeeping genes and was performed on each representative isolate as previously described.16 Each different sequence is assigned a distinct allele (a number) and the combinations of the alleles define the sequence type (ST).
Isolates of STs that were shared across both pigs and chickens were re-examined by rep-PCR.
Analysis of the RIRDC database for P. multocida
The RIRDC MLST database was searched for isolates that had the same ST as the pig isolates and compared. The website is based at the University of Oxford21 and was funded by the Wellcome Trust.
Results
The Heddleston serotyping scheme could not clearly allocate 77% (33/43) of the isolates from pigs, with these isolates being either non-typable (NT) or cross-reacting. All isolates could be assigned an LPS type. Of the eight recognised LPS genotypes, only four were detected in the pig and poultry isolates: L1, L3, L4 and L6 (Supplementary Tables 1, 2). For the pig isolates, the LPS PCR assigned all isolates to an LPS type. There were two poultry isolates that could not be typed by the LPS PCR method. Both of these isolates have been previously reported, one as cross-reacting with serovars 13 and 817 and the other as NT.16
LPS on farms
Some pig farms had more than one LPS type, with isolates of L3 and L6 on the same farm. As an example, Farm 6 had isolates of L3 type that were found to be Heddleston serovars 1/3 cross-reacting, 10 and NT. The L6 isolates were assigned to Heddleston serovar 10 and NT. Similarly, Farm 26 had an L3 isolate shown to serovar 3/4 cross-reacting and an L6 isolate that was serovar 1. Farms 31 and 32 had L3 and L6 isolates, with all being found to be NT. These differences within a farm were also seen in the rep-PCR results, with all of the aforementioned isolates having a different profile, except for the two L3 isolates from Farm 31, which had the same rep-PCR profile.
Rep-PCR
The rep-PCR revealed 20 different profiles for the 43 pig isolates (47% diversity; Supplementary Table 1). The 21 isolates with the LPS L3 genotype displayed 10 different patterns, while the 20 isolates with LPS L6 genotype displayed 8 patterns and the 2 isolates of LPS L1 genotype displayed 2 patterns.
MLST
The 22 pig isolates examined by MLST had 16 STs, with 4 STs (specifically STs 50, 124, 167 and 185) having multiple isolates (Supplementary Table 1). The 41 poultry isolates consisted of 37 different STs, with only STs 7, 8 and 9 having multiple isolates (Supplementary Table 2). Four new STs were recognised in the pig isolates, being ST 326 (a member of clonal complex (CC) ST20), ST 327 (member of CC ST74), ST 328 (member of CC ST13) and ST 329 (member of CC 20).
There were five STs (STs 7, 11, 20, 24 and 58) shared between the poultry and pig isolates (Table 1). The isolates of the shared STs were always the same LPS PCR type within the ST across the two host species. There was some variation within the Heddleston serotyping results. As an example, the ST 7 isolates were found to be serovar 3 (the pig isolate and one poultry isolate) and NT (poultry isolate). ST 11 also had NT for the poultry isolate and the pig isolate had a serovar 4 result. The rest of the shared STs had the same serovar for both pigs and poultry. All of these isolates examined by MLST were either LPS PCR type L1 or L3, which corresponds to serovars 1 and 14 (L1) and serovars 3 and 4 (L3).17 The rep-PCR analysis showed that all three isolates of ST 7 and the two isolates of ST 24 had the same pattern (Table 2). All other isolates of the three shared STs had different rep-PCR patterns, although the difference was only one band for three of the shared STs (Figure 1).
| Host species | Isolate no. | ST | LPS genotype | Heddleston Serotype | Rep-PCR profile | CC |
|---|---|---|---|---|---|---|
| Chicken | 48 | 7 | L3 | 3 | A | |
| Chicken | 1434 | 7 | L3 | NT | A | |
| Pig | 1812 | 7 | L3 | 3 | A | |
| Chicken | 49 | 11 | L1 | NT | B* | |
| Pig | 1463 | 11 | L1 | 4 | C* | |
| Chicken | 1417 | 20 | L3 | 4 | D* | |
| Pig | 1475 | 20 | L3 | 4 | E* | |
| Chicken | 83 | 24 | L3 | NT | F | ST9 |
| Pig | 1676 | 24 | L3 | NT | F | ST9 |
| Chicken | 878 | 58 | L1 | 1, 4 | G* | ST58 |
| Pig | 1732 | 58 | L1 | 1, 4 | H* | ST58 |
- * Isolates of the same ST differ by only one band in the rep-PCR (see Figure 1).
- CC, clonal complex; LPS, lipopolysaccharide; Rep-PCR, repetitive element PCR fingerprinting assay; ST, multilocus sequence typing (MLST) sequence type.
| ST | Host | No. from Australia | No. from other countries |
|---|---|---|---|
| 7 | Avian/porcine | 6 | 0 |
| 11 | Avian/porcine | 2 | 0 |
| 13 | Avian/porcine/bovine/human | 0 | 71 |
| 20 | Avian/porcine/feline | 18 | 0 |
| 24 | Avian/rabbit | 2 | 1 |
| 27 | Avian/porcine | 0 | 16 |
| 50 | Avian/porcine/bovine/rabbit/? | 0 | 26 |
| 58 | Avian/porcine/ovine | 2 | 2 |
| 74 | Avian/porcine/rabbit/human | 0 | 19 |
| 124 | Ovine | 0 | 4 |
| 167 | Porcine | 2 | 0 |
| 185 | ? | 1 | 0 |
| 326 | Porcine | This study | 0 |
| 327 | Porcine | This study | 0 |
| 328 | Porcine | This study | 0 |
| 329 | Porcine | This study | 0 |
- NT, non-typable; RIRDC, Rural Industries Research & Development Corporation; ST, multilocus sequence typing (MLST) sequence type.
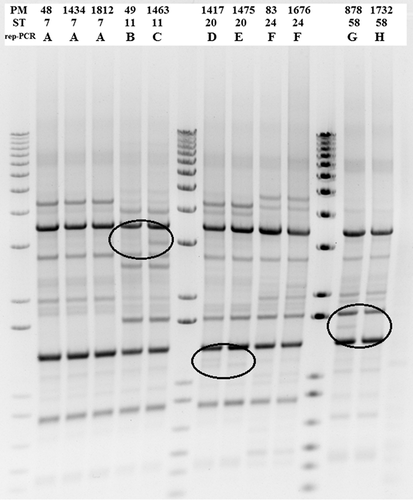
The STs shared across pigs and poultry that were identified in our study (ST7, ST11, ST20, ST24 and ST58) mostly consisted of Australian isolates. The database for the P. multocida MLST (https://pubmlst.org/pmultocida/) reports ST7, ST11 and ST20 as only occurring in Australia, with all of them having been reported from poultry and pigs. ST7 was found in isolates from four chickens, one unknown avian species and one pig, ST11 from one turkey and one pig and ST20 from nine chickens, seven turkeys, one pig and one cat (Table 2). ST24 was reported from two chickens in Australia and one rabbit in Italy and ST58 was reported from one chicken and one pig from Australia, but also from a sheep in New Zealand and a partridge from Belgium (Table 2).
Of the 16 STs observed for the pig isolates of P. multocida, four were new STs and one ST (ST 167) has only been previously recorded from pigs in Australia according to the RIRDC P. multocida MLST database. One isolate of ST 185 had an unknown host, but all other 10 STs had other species listed (Table 2).
Of the 37 STs from the P. multocida isolates from poultry, 24 have only been observed in poultry isolates according to the RIRDC P. multocida MLST database and 13 STs had several host species. The five STs that were found in this study to be shared by both chicken and pig isolates were among the 13 STs listed with different hosts.
Discussion
Two types of serotyping (i.e. capsule and somatic serotyping) are available for serotyping P. multocida. Capsule serotyping of P. multocida recognises 5 distinct capsular groups (A, B, D, E and F) and Heddleston somatic serotyping recognises 16 serovars.1 In the Australian context, Heddleston serotyping has been commonly used for subtyping of P. multocida. The importance of Heddleston serotyping has arisen because it was believed that killed P. multocida vaccines provided protection only against those serovars in the vaccine.10 However, it has been recently shown that vaccines based on killed whole cells of P. multocida only give protection against strains that express highly similar or identical LPS structures.22 As there is inter- and intra-strain LPS structural heterogeneity within an LPS PCR genotype or a serovar,23 it is no longer possible to be confident that isolates of the same serovar or LPS PCR type are indeed cross-protective.
Another problem with Heddleston serotyping is the frequent occurrence of isolates that cannot be serotyped. In the current study, of the 43 pig isolates, a total of 33 were either NT (no reaction with any of the 16 recognised serovars) or gave a non-specific reaction (reaction with > 1 serovar). As well, some of the Heddleston serotyping results did not correlate with the LPS PCR results. As an example, LPS PCR result L3 should be either serovar 3 or serovar 4,17 yet the Heddleston serotyping of these isolates gave results such as NT, 14/15 cross-reacting, 13/15/16 cross-reacting, 4/14 cross-reacting and 1/3 cross-reacting. This lack of correlation was also observed when the LPS PCR was developed17 and in previous application of the PCR.9 In the original development of the PCR,16 the gold standard method of a full chemical and structural analysis showed that LPS PCR was consistently correct, while the traditional serotyping methodology was in error, when these two tests were in disagreement. Hence, the Heddleston serotyping scheme is not suitable for P. multocida, especially for pig isolates.
As was revealed in studies on chicken farms,9 some of the pig farms also had several strains with two LPS genotypes and up to five rep-PCR profiles. These isolates came from diseased pigs and hence it can be assumed that different strains are involved in disease on farms.
The RIRDC MLST scheme was developed with poultry P. multocida isolates.16 Hence, it is not surprising that new STs were found for the pig isolates, as the database lists only 15 Australian pig isolates.
There is evidence that rep-PCR can be more discriminatory than MLST.24 In the current study, the rep-PCR patterns of the isolates from STs that were shared across pig and poultry were either identical (one ST of three isolates and of two isolates) or were nearly identical, as they differed in only one band (two isolates in each of three STs). There is no universally agreed standard for the interpretation of rep-PCR patterns in terms of how many band differences indicate a different ‘strain’.24 In the current study, we adopted the convention that a single band difference meant that isolates were assigned to different patterns. However, in a review of different typing method for P. multocida the suggestion was made that isolates that are indistinguishable or only have minor differences by a variety of typing methods should be classified as genetically related, which means that they are clonal.25 Hence, the overall conclusion is that the isolates from four of the five STs shared between pigs and chickens are example of strains that are shared across these two hosts.
Our finding that some P. multocida STs were shared between the two hosts (poultry and pigs) is in contrast with observations from a study in 201126 looking at bovine P. multocida isolates. That study examined data then available from the RIRDC MLST scheme and showed no relationship of STs between pig and poultry isolates of P. multocida.26 The shared STs identified in our study (ST7, ST11, ST20, ST24 and ST58) mostly consisted of Australian isolates, with only ST58 consisting of isolates from animal species from overseas. It is possible that regional influences play a role in the occurrence of STs. As an example, ovine isolates from Spain and New Zealand have been shown to belong to distinct STs.26
It is worth noting that the RIRDC MLST database currently contains a total of 21 isolates of P. multocida from humans. As shown in Table 3, isolate STs 13 and 74 have been recognised in pigs and poultry as well as humans. This raises the possibility of zoonotic potential of at least some STs. As well, this sharing of genotypes across pigs, poultry and humans raises biosecurity issues.
Conclusion
The fact that the P. multocida isolates from poultry and pigs that shared the same STs had identical or nearly identical rep-PCR patterns, the same LPS genotype and, in most cases, the same Heddleston serovar would indicate that the pig is a potential reservoir for P. multocida for chickens and vice versa. This has implications for biosecurity considerations, including the restriction of visitors and staff that have been in contact with domestic or commercial poultry prior to entering the pig shed. This might also have implication for wild birds, especially water birds on piggery effluent ponds. The same biosecurity precaution would apply with regard to staff or visitor contact with domestic or commercial pigs prior to contact with poultry and the control of wild pigs around poultry farms.
Acknowledgments
This publication made use of the PubMLST website (http://pubmlst.org/) developed by Keith Jolley (Jolley & Maiden 2010, BMC Bioinformatics, 11:595) and sited at the University of Oxford. The development of that website was funded by the Wellcome Trust.
Conflicts of interest and sources of funding
The study was funded by the Australian Poultry CRC Ltd. The authors declare no conflicts of interest for the work presented here.




