Maternal genetic phylogeography analysis of Kyrgyz native cattle
Abstract
This study presents the first mitochondrial DNA analysis of native cattle in Kyrgyzstan, examining sequences from the D-loop region of 20 Kyrgyz native cattle. A phylogenetic tree was constructed to estimate the genetic diversity and lineages, revealing two major haplogroups (T and I). Regional analysis showed a significant frequency difference (p < 0.05) between these haplogroups: Haplogroup I was found to be more prevalent (0.556) in the lower elevation areas (elevation ~1000 m) and less prevalent (0.100) in the higher elevations (elevation ~2500 to 3000 m), suggesting that environmental factors influence genetic distribution among Kyrgyz cattle. Three sub-haplogroups T2, T3, and T4 were observed within major haplogroup T, whereas only one sub-haplogroup (I1) was observed in this study. We also compared the distribution patterns of haplogroups T and I in Kyrgyz with those of some areas in Asia previously reported and found that in Central and Northeast Asia, haplogroup T was dominant, but the gene flow of haplogroup I was also present. The results of this study underscore the importance of regional environmental factors, including altitude, in shaping the genetic structure of livestock populations and would be useful to understand the historical movements and adaptations of Kyrgyz native cattle.
1 INTRODUCTION
It is believed that domestic cattle (Bos taurus) originated from the aurochs (Bos primigenius), with the common ancestor having lived across a wide range of the Eurasian continent and North Africa around the end of the Pleistocene and the early Holocene eras (approximately 11,000 years ago) (Meadow, 1993; van Vuure, 2005). The domestication of taurine cattle (B. taurus) is thought to have first occurred during the Neolithic period (approximately 9000 to 11,000 years ago) in the Fertile Crescent region of the Middle East. Additionally, B. primigenius namadicus, the ancestor of zebu cattle (Bos indicus), inhabited the Indian region and is believed to have been domesticated in the Indus Valley approximately 7000 to 9000 years ago, roughly 2000 years after B. taurus (Achilli et al., 2008; Ajmone-Marsan et al., 2010; Chen et al., 2009; Loftus et al., 1994, 1999; Troy et al., 2001).
Kyrgyzstan is a landlocked country in Central Asia on the Eurasian continent, an important region along the Silk Road that connects the east and west of the continent. Nearly 90% of Kyrgyzstan's territory consists of mountainous areas of <1500 m elevation. Due to its harsh terrain and climate, livestock such as cattle, sheep, and horses are primarily pastured between the mountainous and lowland regions of Kyrgyzstan, while yaks also habit in the higher elevations. Kyrgyzstan is located near the centers of cattle domestication and along the Silk Road's Oasis Road route. Therefore, the genetic information of cattle in this region is important for understanding migration from the centers of domestication into Asia.
This study's analysis of Kyrgyz native cattle, which leverages mitochondrial DNA (mtDNA) control-region sequences, not only illuminates the genetic structure of cattle within Kyrgyzstan but also enhances our understanding of cattle migration patterns across Central Asia. Our result would contribute to a better understanding of the genetic history of livestock along the Silk Road and Kyrgyzstan's role in the history of animal husbandry and trade.
2 MATERIALS AND METHODS
Genomic DNA was extracted from the blood samples of unrelated 20 native cattle (4 males and 16 females), which sampled randomly in August 2019, from three regions in Kyrgyzstan: Son-Kul, Naryn Province (4 males and 6 females), Tokmok, Chui Province (9 females), and Issyk-Kul, Karakol Province (1 female) (Figure 1a and Table S1). All Kyrgyz native cattle reveal B. taurus morphology with humpless (Figure 1b,c). The mtDNA D-loop region (907–910 bp) was amplified and sequenced according to the method described previously (Lin et al., 2013; Yamanaka et al., 2019). Following sequencing, we conducted phylogenetic analyses using the maximum likelihood (ML) method. The analyses were based on the genetic distances and used the reference sequences LC013968 (T1), AB117049 (T2), V00654 (T3), LC013966 (T4), AB268579 (I1), and AB268559 (I2). The phylogenetic relationships were estimated by performing 100 bootstrap replications.
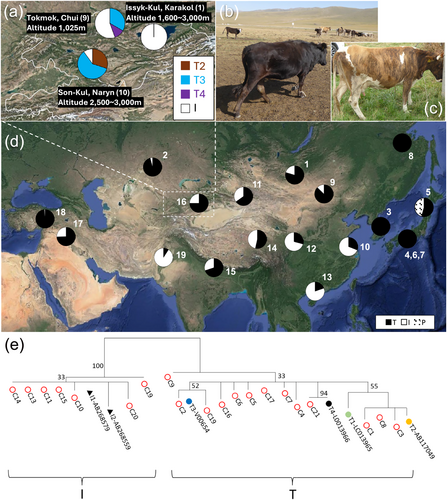
The distribution of cattle mtDNA haplogroups across the Eurasian continent was investigated. In addition to the results of this study, data from 2064 individuals concerning the mtDNA D-loop region (910 bp) of domestic cattle from previous studies were used to examine the distribution frequency from the centers of domestication to East Asia (Figure 1d and Table S2). Fisher's exact test was performed for testing significant haplogroup frequency between two regions of Kyrgyzstan.
3 RESULTS AND DISCUSSION
This is the first report to conduct mtDNA analysis of cattle in Kyrgyzstan. We obtained the sequences of the mtDNA D-loop region (907–910 bp) from 20 Kyrgyz native cattle and deposited the sequences in the DDBJ database (accession numbers: LC810626–LC810645). We constructed a phylogenetic tree of Kyrgyz cattle using representative mtDNA D-loop sequences of cattle (Figure 1e). The phylogenetic tree indicated that Kyrgyz cattle have two major haplogroups: T and I. All haplotypes are previously observed in other countries (mainly in China, Kazakhstan, and India).
The distribution of the haplogroups and sub-haplogroups in three regions of Kyrgyzstan is presented in Figure 1a. The frequency of haplogroup I varies regionally within Kyrgyzstan, being 10.0% in Son-Kul, Naryn and 55.6% in Tokmok, Chui. This frequency difference is significant (p = 0.0495) and is probably caused by regional elevation. Tokmok, Chui, with an elevation of about 1000 m and a low average temperature (annual average: 9.5°C, monthly averages: −10°C to 30°C), is considered a suitable environment for both B. taurus and B. indicus. Conversely, Son-Kul, Naryn, located at higher elevations of 2500–3000 m, has an even lower annual average temperature of −3.5°C (monthly averages: −14.8°C to 17.8°C) is suitable for B. taurus due to its adaptability to colder regions.
Previous research has revealed that B. taurus mtDNA can be largely classified into six sub-haplogroups: T and T1–T5. Sub-haplogroup T1 is observed mainly in Africa, while haplogroups T and T2, T3, T4, and T5 are found in the Middle East/Near East, Europe, East Asia, and Italy, respectively (Baig et al., 2005; Lei et al., 2006; Magee et al., 2007). Our phylogenetic tree indicated that there are three sub-haplogroups (T2, T3, and T4) within Kyrgyzstan (Son-Kul, Naryn, T2: n = 3, T3: n = 6 and Tokmok, Chui, T3: n = 3, T4: n = 1) (Figure 1a and Table S1). Meanwhile, all animals in haplogroup I were found to belong to sub-haplogroup I1. Sub-haplogroup T4 is observed exclusively in East Asia, spanning from Kyrgyzstan through Northern and Central China along the Silk Road, while it is not observed west of the Fertile Crescent (Jia et al., 2010; Lei et al., 2006; Mannen et al., 1998, 2004, 2017; Troy et al., 2001; Xia et al., 2019). Currently, the westernmost locations of haplogroup T4 are in Kazakhstan (Yamanaka et al., 2019) and Kyrgyzstan (this study). The finding of haplogroup T4 in Kyrgyzstan demonstrates the importance of the Silk Road, which facilitated the trade of materials, including livestock. This distribution pattern and frequency are crucial for understanding the origins and spread of the East Asia-specific T4 sub-haplogroup.
The distribution frequencies of haplogroups T and I across Asia, particularly from the centers of domestication to the East Asian region, reveal that haplogroup T is dominant in Central and Northeast Asia (Figure 1d and Table S2). This pattern suggests that populations carrying haplogroup T spread to East Asia via the Oasis Road as early domesticated animals. Archaeological and molecular evidence has shown that B. taurus first appeared in North China in the late Neolithic period between approximately 5000 and 3000 years ago (Cai et al., 2014; Mannen et al., 2020). Moreover, Verdugo et al. (2019) performed genome-wide analysis of ancient Near Eastern B. taurus DNA samples and revealed the rapid and widespread introgression of B. indicus from the Indus Valley during the Bronze Age (approximately 4200 years ago). They also showed that the introgression was mainly by the arid-adapted B. indicus male. Taking these results into consideration with the results of the current study, the expansion of the B. indicus type (mitochondrial haplogroup I) into Central Asia may have occurred much later; for example, by the expanding Mongol Empire in the 13th century (Mannen et al., 1998). In conclusion, the mtDNA information of Kyrgyz native cattle contributes to our understanding of the process of northern cattle spread to East Asia.
ACKNOWLEDGMENTS
This work was supported financially by a Grant-in-Aid for Scientific Research (B) (Grant No. 17H04637 and 21H02343) by JSPS. We thank the members of the Society for Researches on Native Livestock for collecting DNA samples.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.




