Whole-genome resequencing to identify candidate genes for the QTL for oleic acid percentage in Japanese Black cattle
Abstract
In our previous study, we detected a QTL for the oleic acid percentage (C18:1) on BTA9 in Japanese Black cattle through a genome-wide association study (GWAS). In this study, we performed whole-genome resequencing on eight animals with higher and lower C18:1 to identify candidate polymorphisms for the QTL. A total of 39,658 polymorphisms were detected in the candidate region, which were narrowed to 1993 polymorphisms within 23 genes based on allele differences between the high and low C18:1 groups. We subsequently selected three candidate genes, that is, CYB5R4, MED23, and VNN1, among the 23 genes based on their function in fatty acid metabolism. In each candidate gene, three SNPs, that is, CYB5R4 c.*349G > T, MED23 c.3700G > A, and VNN1 c.197C > T, were selected as candidate SNPs to verify their effect on C18:1 in a Japanese Black cattle population (n = 889). The statistical analysis showed that these SNPs were significantly associated with C18:1 (p < 0.05), suggesting that they were candidates for the QTL. In conclusion, we successfully narrowed the candidates for the QTL by detecting possible polymorphisms located within the candidate region. It is expected that the responsible polymorphism can be identified by demonstrating their effect on the gene's function.
1 INTRODUCTION
In our previous study, we identified a QTL for the oleic acid percentage (C18:1) on BTA9 when a genome-wide association study (GWAS) was conducted on a Japanese Black cattle population (Kawaguchi et al., 2018). We searched for genes involved in fatty acid metabolism using the NCBI database to identify candidate genes around the QTL. We ultimately selected VNN1 as a candidate gene because it was involved in fatty acid synthesis. A putative candidate SNP, c.197C > T (T66M), was detected within the gene. To verify the effect of this SNP on C18:1, we investigated the association between this SNP and C18:1 in a Japanese Black cattle population. Although the SNP was significantly associated with C18:1, its p-value was higher than that of the most significant SNP identified during the GWAS. Therefore, we concluded that another polymorphism might be responsible for the QTL.
Whole-genome resequencing has recently been used for detecting polymorphisms that might affect the economic traits of livestock animals. Jiang et al. (2016) used whole-genome resequencing to detect short indels in eight dairy cattle with high and low estimated breeding values (EBV) for their milk composition traits. They compared genotypes in eight animals to select candidate indels. As the responsible indel would show different genotype between high EBV and low EBV animals, they selected indels showing almost the same genotype in four animals in each group and showing the different alleles between high and low animals. To select candidate genes from several genes containing the candidate indels, they collected some information on the pathways, QTL registered on databases, and positions of the significant SNPs identified by GWAS in previous reports. They ultimately identified 11 candidate genes based on functional and positional information. Li et al. (2015) also used whole-genome resequencing in 10 birds to detect SNPs for pH value of chicken meat in a fine-mapped QTL on chromosome 1. Half of the birds were homozygous for the high-QTL allele (QQ group), and the remaining birds were homozygous for the low QTL allele (qq group). They extracted SNPs that exhibited different alleles between the high and low QTL groups and focused on the SNPs expected to affect the gene's function, according to their annotations (such as non-synonymous, UTR, splicing sites, CpG island, and promoter regions). They conclusively identified 129 SNPs as candidates for the QTL.
Previous studies suggest that whole-genome resequencing is a useful method for identifying candidate polymorphisms and genes. The objective of this study was to identify candidate polymorphisms and genes for the QTL for C18:1 on BTA9 using whole-genome resequencing.
2 MATERIALS AND METHODS
2.1 Sample selection
In our previous study, we used Japanese Black cattle population comprising 1,836 animals that had been bred in the Hyogo Prefecture (Kawaguchi et al., 2018). After correcting for the C18:1 phenotype of each animal using an analytical model, we selected a total of 200 animals for conducting pool-based GWAS: 100 of these animals had higher C18:1 and the other 100 had lower C18:1 in the previous study.
In this study, eight animals were selected from the 200 animals used in the GWAS for whole-genome resequencing based on their sires and genotype of the most significant SNP in the GWAS (Hapmap43702-BTA-84086) (Table 1). We selected four animals with the TT genotype from the 100 “high” animals and four animals with the CC genotype from the 100 “low” animals, which were the progenies of different sires among the four animals in the high and low groups. Genomic DNA was extracted from each 50-mg longissimus cervicis muscle sample following the standard phenol–chloroform method.
| Group | Sample | Genotype | Sire | C18:1 | Corrected C18:1 |
|---|---|---|---|---|---|
| High | 1 | TT | 1 | 63.05 | 58.44 |
| 2 | TT | 2 | 59.81 | 56.57 | |
| 3 | TT | 3 | 59.25 | 56.13 | |
| 4 | TT | 4 | 57.89 | 56.37 | |
| Low | 5 | CC | 3 | 49.72 | 50.76 |
| 6 | CC | 5 | 48.39 | 48.79 | |
| 7 | CC | 6 | 47.60 | 49.46 | |
| 8 | CC | 7 | 46.17 | 49.75 |
- Note. Genotype: the genotype of Hapmap43702-BTA-84086, which was the most significantly associated with C18:1 in the GWAS (Kawaguchi et al., 2018).
- Corrected C18:1: the corrected values of C18:1 using an analytical model.
2.2 Whole-genome resequencing
DNA degradation was monitored based on its concentration by spectrometry, fluorometry, and 1% agarose gel electrophoresis. A paired-end library was constructed using high-quality DNA for each individual, and its read length was 150 bp. Sequencing was performed using a HiSeq X Five Sequencing System (Illumina Inc., San Diego, CA, USA). Sequencing data were normalized by Genedata Expressionist 9.1.4a. We mapped the reads to the cattle reference genome assembly (UCSC bosTau8) downloaded from the UCSC Genome Browser assembly (https://genome-asia.ucsc.edu/cgi-bin/hgGateway) using BWA-MEM 0.7.12. and excluded PCR duplicates using Picard 2.2.4. GATK 3.6 (2016-12-08-g1c2527f) was used to call polymorphisms by comparing the genome sequences, including the reference sequence. The polymorphisms were annotated to the gene reference (NCBI RefSeq) based on their location (intron, exon, untranslated region, upstream, downstream, splice site, and intergenic region) and characteristics (synonymous/non-synonymous amino acid replacement, gain/loss of start/stop site, and frameshift mutations) using SnpEff v4.2 with the reference sequence (bosTau8).
2.3 Narrowing the polymorphisms and genes
In our previous study, we determined the region 5 Mbp upstream and downstream of Hapmap43702-BTA-84086 (64.9 –74.9 Mbp) to be the candidate region (Kawaguchi et al., 2018). In this study, we selected polymorphisms in the candidate region from among all the polymorphisms detected by whole-genome resequencing.
We first excluded intergenic polymorphisms and subsequently focused on the linkage disequilibrium (LD) between the polymorphisms and Hapmap43702-BTA-84086 because the responsible polymorphism is in LD with Hapmap43702-BTA-84086. We compared genotypes of four animals in high group with those of four animals in low group as an indicator of LD. The Hapmap43702-BTA-84086 genotypes completely differed between the high and low groups; it was defined that there were a total of eight allele differences between the high and low groups. Therefore, polymorphisms with a large number of allele differences were expected to be in LD with the SNP. In this study, we focused on the polymorphisms with over three allele differences.
We investigated the function of genes containing these polymorphisms in the NCBI database and previous reports and determined candidate genes based on their function in fatty acid metabolism, such as synthesis, transport, desaturation, and oxidation.
2.4 Genotyping the candidate SNPs
Polymorphisms were prioritized as candidates based on their location and characteristics. We selected candidate polymorphisms that were the most likely to affect the function of each candidate gene.
To test the effect of candidate polymorphisms on C18:1, we used a Japanese Black cattle population (n = 899) that had been randomly selected from 1,836 individuals bred in the Hyogo Prefecture, Japan, and graded them from 2010 to 2012. This population contained at least 10 offspring per each sire. We genotyped CYB5R4 c.*349G > T and MED23 c.3700G > A by PCR-RFLP. The primer sets for PCR amplification were designed based on the GenBank sequence (AC_000166.1) using OLIGO 7.41. The PCR reaction was performed at the annealing temperature and extension period shown in Table 2. The PCR products for CYB5R4 c.*349G > T and MED23 c.3700G > A were digested using MslI and ScaI, respectively.
| Polymorphism | Primer set | AT (°C) | EP (s) | PL (bp) | RE |
|---|---|---|---|---|---|
| CYB5R4c.*349G | F: 5′- AGA CAG CAC ATA CTA ACT GGG A -3′ | 60 | 60 | 908 | MslI |
| R: 5′- GAG GCT ACA CTT CCA TTA TCT T -3′ | |||||
| MED23c.3700G > A | F: 5′- TGT ACT GCA TAC CTG ATA GTA GC -3′ | 62 | 30 | 695 | ScaI |
| R: 5′- CAT CCC ACA CCA AAC TCA TAG GC -3′ |
- Note. AT, annealing temperature; EP, extension period; PL, PCR product length; RE, restriction enzyme.
2.5 Statistical analysis
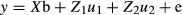
The e-values of 899 animals were analyzed by one-way analysis of variance (ANOVA) with genotype as a source of variation and Tukey-Kramer's honestly significant difference (HSD) test implemented in JMP13 (SAS institute Inc., Cary, NC, USA).
The proportion of additive genetic variance was calculated following the method of Matsuhashi et al. (2011). LD coefficients (r2) between the polymorphisms were calculated using HAPLOVIEW 4.0.
3 RESULTS
3.1 Detection of candidate polymorphisms by genome resequencing
We conducted whole-genome resequencing in eight Japanese Black cattle (Supporting Information Table S1). A total of 39,658 polymorphisms were identified in the candidate region (Chr 9: 64,956,436–74,956,436) by comparing genome sequences between nine animals, including the reference (bosTau8). Of these, 10,045 polymorphisms were located within the genes. We further narrowed the polymorphisms to 1993 with over three allele differences between the high and low groups, which were located within a total of 23 genes. These SNPs were counted based on genes and annotations to select the candidate polymorphisms (Supporting Information Table S2).
3.2 Candidate genes in terms of gene function
We investigated the functions of the 23 genes based on information in the NCBI database and previous reports (Supporting Information Table S3). Three genes, that is, CYB5R4, MED23, and VNN1, were determined to be candidate genes in terms of their function in fatty acid metabolism.
CYB5R4 is an electron donor for fatty acid desaturation by stearoyl-CoA desaturase (SCD) (Deng et al., 2010; Zhu et al., 2004). Larade et al. (2008) reported that the C18 desaturation index (C18:1/C18:0) was markedly lower in CYB5R4-knockout mouse than in a wild-type mouse. Polymorphisms within the CYB5R4 gene may affect SCD activity, which would alter the ratio between C18:1 and C18:0.
MED23 is a subunit of the Mediator complex and transmits information between RNA polymerase II and transcription factors, such as FOXO1 (Chu et al., 2014; Knuesel & Taatjes, 2011). A previous study reported that FOXO1 suppresses the expression of acetyl-CoA carboxylase (ACC), which catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, that is, the first step of long-chain fatty acid synthesis (Bastie et al., 2005). MED23 may control fatty acid synthesis by regulating the expression of ACC.
VNN1 is an enzyme that controls the amount of pantethine generated by pantetheinase activity (Kavian et al., 2015; Pitari et al., 2000). Acetyl-CoA and malonyl-CoA are converted into acetyl-ACP and malonyl-ACP, respectively, during one step of fatty acid synthesis. As pantethine bypasses this reaction as a metabolically active substrate for CoA and ACP (Kelly, 1997), VNN1 appears to be involved in fatty acid synthesis.
3.3 The effect of the candidate SNPs on C18:1
A total of 77, 125, and 28 polymorphisms were detected within the CYB5R4, MED23, and VNN1 genes, respectively (Supporting Information Table S2). We selected one candidate polymorphism from each candidate gene to test its effect on C18:1. We primarily focused on amino acid substitutions and secondarily focused on SNPs related to gene expression (5′UTR, 3′UTR, the promoter region, and splice site). The amino acid substitutions, V1234I and T66M, were selected within MED23 and VNN1 genes, respectively, while we selected the SNP on 3′UTR in CYB5R4 gene as amino acid substitution was not detected within the gene (Supporting Information Table S2). In short, CYB5R4 c.*349G > T, MED23 c.3700G > A (V1234I), and VNN1 c.197C > T (T66M) were selected as candidate SNPs for the QTL. VNN1 c.197C > T was selected as a candidate SNP in our previous study (Kawaguchi et al., 2018).
We previously genotyped the most significant SNP in GWAS (Hapmap43702-BTA-84086) and VNN1 c.197C > T for a Japanese Black cattle population (n = 899) (Kawaguchi et al., 2018). In addition, we genotyped CYB5R4 c.*349G>T and MED23 c.3700G>A for the same population in this study. The minor allele frequencies of Hapmap43702-BTA-84086, CYB5R4 c.*349G>T, MED23 c.3700G > A, and VNN1 c.197C > T were 0.261, 0.272, 0.280, and 0.201, respectively (Table 3). These SNPs were in relatively high LD with Hapmap43702-BTA-84086 (r2 = 0.53–0.83) (Fig. 1). In our previous study, statistical analysis revealed that Hapmap43702-BTA-84086 and VNN1 c.197C > T were significantly associated with C18:1 (p = 0.0080 and 0.0162, respectively) (Kawaguchi et al., 2018). Therefore, we also conducted statistical analysis for CYB5R4 c.*349G > T and MED23 c.3700G > A to investigate and compare their effects on C18:1. ANOVA revealed that both SNPs were significantly associated with C18:1 (p = 0.0075 and 0.0295) (Table 4). Significant differences between genotypes were observed by the Tukey–Kramer's HSD test. The proportions of additive genetic variance for Hapmap43702-BTA-84086, CYB5R4 c.*349G > T, MED23 c.3700G > A, and VNN1 c.197C > T were 3.70, 4.20, 3.30, and 2.24, respectively.
| Polymorphism | Position | Genotype frequency (n) | Allele frequency | |||
|---|---|---|---|---|---|---|
| Hapmap43702-BTA-84086 | 69,956,436 | TT | TC | CC | T | C |
| 0.528 (475) | 0.420 (378) | 0.051 (46) | 0.739 | 0.261 | ||
| CYB5R4c.*349G > T | 66,377,383 | GG | GT | TT | G | T |
| 0.505 (454) | 0.446 (401) | 0.049 (44) | 0.728 | 0.272 | ||
| MED23 c.3700G > A | 70,521,413 | AA | AG | GG | A | G |
| 0.491 (441) | 0.458 (412) | 0.051 (46) | 0.720 | 0.280 | ||
| VNN1 c.197C > T | 71,851,690 | CC | CT | TT | C | T |
| 0.630 (566) | 0.338 (304) | 0.032 (29) | 0.799 | 0.201 | ||
- Note. The results of Hapmap43702-BTA-84086 and VNN1 c.197C > T were cited from Kawaguchi et al. (2018).

| Polymorphism | e-values | p-value | %VA | ||
|---|---|---|---|---|---|
| ±SE | |||||
| Hapmap43702-BTA-84086 | TT | TC | CC | 0.0080 | 3.70 |
| 0.032a | −0.234a,b | −0.736b | |||
| ±0.085 | ±0.095 | ±0.273 | |||
| CYB5R4c.*349G>T | GG | GT | TT | 0.0075 | 4.20 |
| 0.056a | −0.261b | −0.636b | |||
| ±0.087 | ±0.093 | ±0.279 | |||
| MED23 c.3700G>A | AA | AG | GG | 0.0295 | 3.30 |
| 0.018 | −0.205 | −0.652 | |||
| ±0.088 | ±0.091 | ±0.274 | |||
| VNN1 c.197C>T | CC | CT | TT | 0.0162 | 2.24 |
| −0.050a | −0.158a | −1.054b | |||
| ±0.078 | ±0.106 | ±0.344 | |||
- Note. The results of Hapmap43702-BTA-84086 and VNN1 c.197C > T were cited from Kawaguchi et al. (2018).
- e-values: the mean of e-values for C18:1 for each genotype.
- %VA: the proportion of additive genetic variance.
- a,bMeans with different superscript are significantly different between genotypes.
4 DISCUSSION
We focused on three genes, that is, CYB5R4, MED23, and VNN1, as candidates for the QTL based on whole-genome resequencing data and functional information. For each gene, we selected CYB5R4 c.*349G > T, MED23 c.3700G > A, and VNN1 c.197C > T as candidate SNPs. One of them, VNN1 c.197C > T, has been selected as the candidate SNP and genotyped in a Japanese Black cattle population (n = 899) in our previous study (Kawaguchi et al., 2018). We additionally genotyped the other two SNPs in the same Japanese Black cattle population and then calculated the LD coefficient between the SNPs and Hapmap43702-BTA-84086. Previous studies reported that the average LD coefficient (r2) between markers was <0.1 in Japanese Black cattle, even at a distance of 2 Mbp (McKay et al., 2007), and the LD block size ranges were 0.022–2.5 Mbp in three chromosomes in Japanese Black cattle (Watanabe et al., 2008). However, the population in our study was expected to have a wider LD block as they undergo uniquely closed breeding (Nakajima et al., 2018). In this study, CYB5R4 c.*349G > T exhibited a high LD coefficient with Hapmap43702-BTA-84086 (r2 = 0.77) despite its location at approximately 4 Mbp away from the SNP. This result is consistent with the expectation of a longer average distance between polymorphisms in LD of the population compared with that in other Japanese Black cattle populations. In addition, all three candidate SNPs were in relatively high LD, suggesting that the number of allele differences is an appropriate indicator of LD with Hapmap43702-BTA-84086.
In this study, we compared the effect of three candidate SNPs on C18:1 in the Japanese Black cattle population. Although three SNPs were significantly associated with C18:1, only CYB5R4 c.*349G > T exhibited a smaller p-value than that Hapmap43702-BTA-84086. In addition, this SNP exhibited the highest additive genetic variance among the four SNPs, including Hapmap43702-BTA-84086. These results suggest that CYB5R4 c.*349G > T is the most likely candidate for the QTL among the three candidate SNPs, which were located on CYB5R4 3′UTR, and may, therefore, affect gene expression. CYB5R4 is an essential electron donor for Δ9 fatty acid desaturation by SCD (Holloway & Katz, 1972; Larade et al., 2008; Zhu, Qiu, Yoon, Huang, & Bunn, 1999), and the SNP may be responsible for C18:1 through the alteration of SCD activity based on the CYB5R4 gene expression level.
Although their p-values were larger than that of CYB5R4 c.*349G > T, the other two SNPs exhibited a similar p-value to that of Hapmap43702-BTA-84086, suggesting that they could also be candidates for the QTL. While we successfully narrowed the candidate polymorphisms and genes based on whole-genome resequencing data and functional information, it was not possible to determine the responsible polymorphism for the QTL in this study. Therefore, we must verify possibilities for all polymorphisms, including CYB5R4 c.*349G > T, MED23 c.3700G > A, and VNN1 c.197C > T, to be the responsible polymorphism. In addition, we must demonstrate the effect of the candidate polymorphisms on the gene's functioning to identify which is responsible. In conclusion, we obtained beneficial information to prioritize candidates in this study. We will identify the responsible polymorphism by conducting further analysis.
ACKNOWLEDGMENTS
We thank Wagyu Registry Association for providing the pedigree information of Japanese Black. This work was supported in part by JSPS KAKENHI Grant Numbers 16H05015.




